The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The goal of vsp is to enable fast, spectral estimation
of latent factors in random dot product graphs. Under mild assumptions,
the vsp estimator is consistent for (degree-corrected)
stochastic blockmodels, (degree-corrected) mixed-membership stochastic
blockmodels, and degree-corrected overlapping stochastic
blockmodels.
More generally, the vsp estimator is consistent for
random dot product graphs that can be written in the form
E(A) = Z B Y^Twhere Z and Y satisfy the varimax
assumptions of [1]. vsp works on directed and undirected
graphs, and on weighted and unweighted graphs. Note that
vsp is a semi-parametric estimator.
You can install the released version of vsp from CRAN
with
install.packages("vsp")You can install the development version of vsp with:
install.packages("devtools")
devtools::install_github("RoheLab/vsp")Obtaining estimates from vsp is straightforward. We
recommend representing networks as igraph objects or sparse
adjacency matrices using the Matrix
package. Once you have your network in one of these formats, you can get
estimates by calling the vsp() function. The result is a
vsp_fa S3 object.
Here we demonstrate vsp usage on an igraph
object, using the enron network from
igraphdata package to demonstrate this functionality. First
we peak at the graph:
library(igraph)
data(enron, package = "igraphdata")
image(sign(get.adjacency(enron, sparse = FALSE)))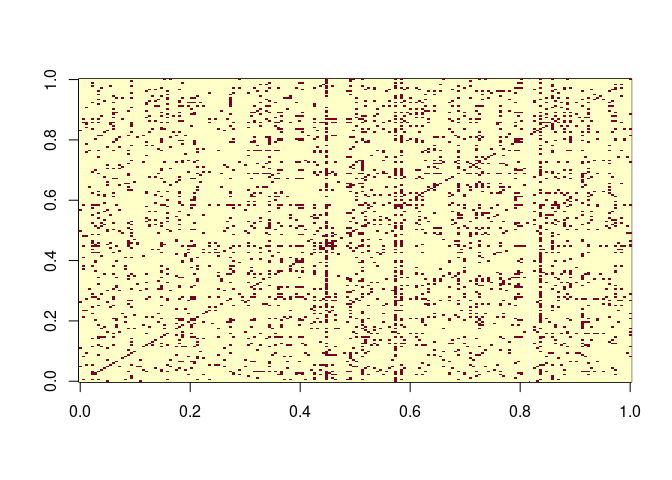
Now we estimate:
library(vsp)
fa <- vsp(enron, rank = 30)
fa
#> Vintage Sparse PCA Factor Analysis
#>
#> Rows (n): 184
#> Cols (d): 184
#> Factors (rank): 30
#> Lambda[rank]: 0.2077
#> Components
#>
#> Z: 184 x 30 [matrix]
#> B: 30 x 30 [matrix]
#> Y: 184 x 30 [matrix]
#> u: 184 x 30 [matrix]
#> d: 30 [numeric]
#> v: 184 x 30 [matrix]get_varimax_z(fa)
#> # A tibble: 184 × 31
#> id z01 z02 z03 z04 z05 z06 z07 z08
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 row0… 2.42e-4 -0.00245 -2.99e-2 3.37e-4 9.96e-5 -0.0114 -0.00849 0.502
#> 2 row0… -2.52e-3 0.00135 6.70e-4 -1.63e-1 -1.47e-2 0.0471 0.190 0.00181
#> 3 row0… 2.98e-4 -0.100 1.17e-4 -3.62e-3 -2.06e-2 0.187 -0.158 0.00303
#> 4 row0… -7.75e-5 -0.0183 1.17e-4 5.42e-2 -5.58e-3 0.00165 -0.0367 -0.00106
#> 5 row0… -2.31e-3 0.00150 2.57e-1 -1.42e-2 -4.38e-2 0.00629 1.18 -0.0179
#> 6 row0… -3.46e-2 -0.0527 -2.61e-2 -1.26e-2 -1.83e-2 0.0282 0.408 -0.0286
#> 7 row0… -1.08e-3 -0.327 -6.01e-1 -6.98e-2 -9.85e-2 -0.0709 0.509 0.0511
#> 8 row0… 1.58e-2 -0.0518 -1.34e-2 -1.03e-2 -4.12e-3 -0.0139 0.225 -0.0244
#> 9 row0… 2.22e-3 0.0752 3.30e-2 -6.50e-4 -5.00e-1 -0.0278 -0.0740 -0.00556
#> 10 row0… 7.13e-4 -0.0119 1.95e-2 -5.06e-3 -7.08e-3 0.00341 -0.00369 13.4
#> # ℹ 174 more rows
#> # ℹ 22 more variables: z09 <dbl>, z10 <dbl>, z11 <dbl>, z12 <dbl>, z13 <dbl>,
#> # z14 <dbl>, z15 <dbl>, z16 <dbl>, z17 <dbl>, z18 <dbl>, z19 <dbl>,
#> # z20 <dbl>, z21 <dbl>, z22 <dbl>, z23 <dbl>, z24 <dbl>, z25 <dbl>,
#> # z26 <dbl>, z27 <dbl>, z28 <dbl>, z29 <dbl>, z30 <dbl>To visualize a screeplot of the singular value, use:
screeplot(fa)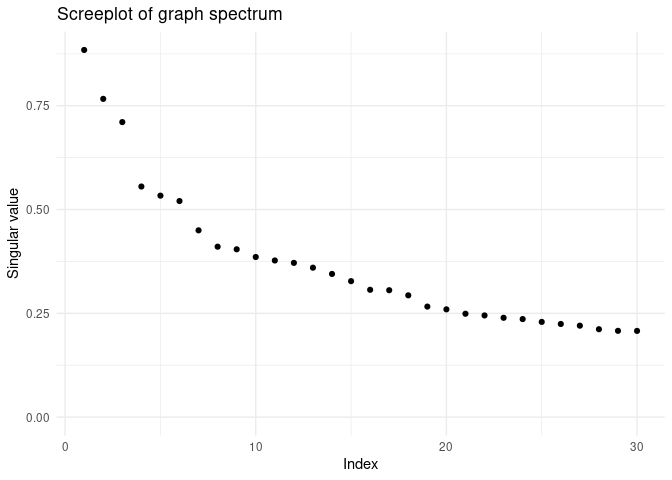
At the moment, we also enjoy using pairs plots of the factors as a diagnostic measure:
plot_varimax_z_pairs(fa, 1:5)
plot_varimax_y_pairs(fa, 1:5)
Similarly, an IPR pairs plot can be a good way to check for singular vector localization (and thus overfitting!).
plot_ipr_pairs(fa)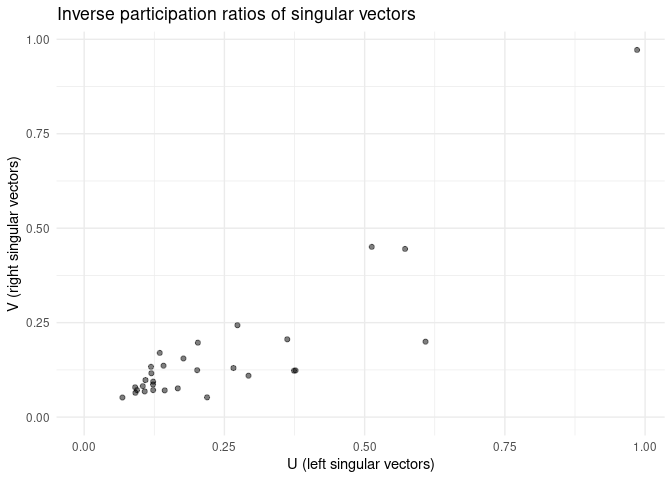
plot_mixing_matrix(fa)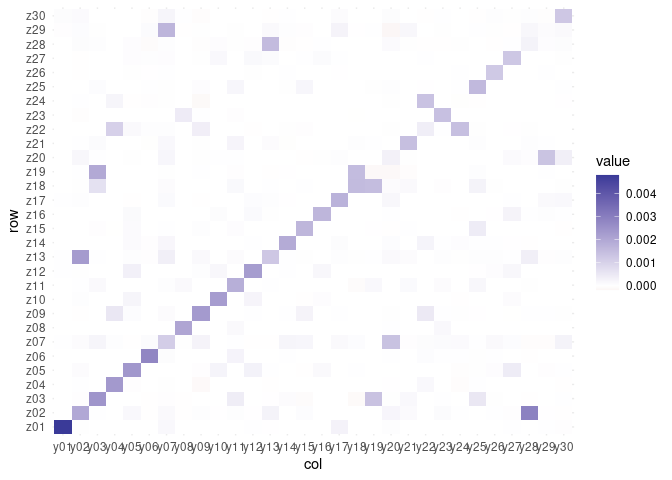
[1] Rohe, Karl, and Muzhe Zeng. “Vintage Factor Analysis with Varimax Performs Statistical Inference.” Journal of the Royal Statistical Society Series B: Statistical Methodology 85, no. 4 (September 29, 2023): 1037–60. https://doi.org/10.1093/jrsssb/qkad029.
Code to reproduce the results from the paper is available here.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.