The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The goal of vimixr is to perform collapsed Variational Inference for DPMM using adaptive inference on the DP concentration parameter as well as covariance hyper-parameter of DP base distribution.
You can install the development version of vimixr from GitHub with:
# install.packages("devtools")
devtools::install_github("annesh07/vimixr")library(vimixr)Let’s generate some toy data. Here the data contains N = 100 samples, D = 2 dimensions and K = 2 clusters.
X <- rbind(matrix(rnorm(100, m=0, sd=0.5), ncol=2),
matrix(rnorm(100, m=3, sd=0.5), ncol=2))In order to obtain the clusters present in X, we apply function from vimixr package that corresponds to a simple case with cluster-inspecific fixed diagonal covariance for the data.
# Fixed-diagonal variance
res <- cvi_npmm(X, variational_params = 20, prior_shape_alpha = 0.001,
prior_rate_alpha = 0.001, post_shape_alpha = 0.001,
post_rate_alpha = 0.001, prior_mean_eta = matrix(0, 1, ncol(X)),
post_mean_eta = matrix(0.001, 20, ncol(X)),
log_prob_matrix = t(apply(matrix(0.001, nrow(X), 20), 1,
function(x){x/sum(x)})), maxit = 1000,
fixed_variance = TRUE, covariance_type = "diagonal",
prior_precision_scalar_eta = 0.001,
post_precision_scalar_eta = matrix(0.001, 20, 1),
cov_data = diag(ncol(X)))
summary(res)
#> Length Class Mode
#> posterior 5 -none- list
#> optimisation 3 -none- list
#> PCA_viz 1 ggplot2::ggplot S4
#> ELBO_viz 1 ggplot2::ggplot S4
#> Seed_used 1 -none- character
plot(res)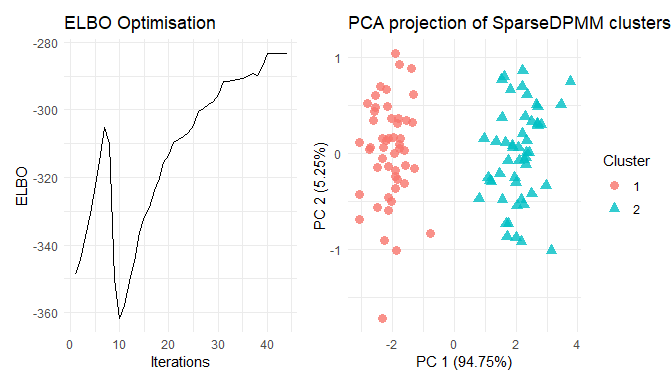
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.