tidyfast v0.4.0

The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
tidyfast v0.4.0

Note: The expansion of dtplyr has made some of
the functionality in tidyfast redundant. See
dtplyr for a list of functions that are handled within that
framework.
The goal of tidyfast is to provide fast and efficient
alternatives to some tidyr (and a few dplyr)
functions using data.table under the hood. Each have the
prefix of dt_ to allow for autocomplete in IDEs such as
RStudio. These should compliment some of the current functionality in
dtplyr (but notably does not use the lazy_dt()
framework of dtplyr). This package imports
data.table and cpp11 (no other
dependencies).
These are, in essence, translations from a more
tidyverse grammar to data.table. Most
functions herein are in places where, in my opinion, the
data.table syntax is not obvious or clear. As such, these
functions can translate a simple function call into the fast, efficient,
and concise syntax of data.table.
The current functions include:
Nesting and unnesting (similar to
dplyr::group_nest() and tidyr::unnest()):
dt_nest() for nesting data tablesdt_unnest() for unnesting data tablesdt_hoist() for unnesting vectors in a list-column in a
data tablePivoting (similar to
tidyr::pivot_longer() and
tidyr::pivot_wider())
dt_pivot_longer() for fast pivoting using
data.table::melt()dt_pivot_wider() for fast pivoting using
data.table::dcast()If Else (similar to
dplyr::case_when()):
dt_case_when() for dplyr::case_when()
syntax with the speed of data.table::fifelse()Fill (similar to tidyr::fill())
dt_fill() for filling NA values with
values before it, after it, or both. This can be done by a grouping
variable (e.g. fill in NA values with values within an
individual).Count and Uncount (similar to
tidyr::uncount() and dplyr::count())
dt_count() for fast counting by group(s)dt_uncount() for creating full data from a count
tableSeparate (similar to
tidyr::separate())
dt_separate() for splitting a single column into
multiple based on a match within the column (e.g., column with values
like “A.B” could be split into two columns by using the period as the
separator where column 1 would have “A” and 2 would have “B”). It is
built on data.table::tstrsplit(). This is not well tested
yet and lacks some functionality of tidyr::separate().Adjust data.table print options
dt_print_options() for adjusting the options for
print.data.table()tidyfast attempts to convert syntax from
tidyr with its accompanying grammar to
data.table function calls. As such, we have tried to
maintain the tidyr syntax as closely as possible without
hurting speed and efficiency. Some more advanced use cases in
tidyr may not translate yet. We try to be transparent about
the shortcomings in syntax and behavior where known.
Each function that takes data (labeled as dt_ in the
package docs) as its first argument automatically coerces it to a data
table with as.data.table() if it isn’t already a data
table. Each of these functions will return a data table.
You can install the stable version from CRAN with:
install.packages("tidyfast")or you can install the development version from GitHub with:
# install.packages("remotes")
remotes::install_github("TysonStanley/tidyfast")#> ℹ Loading tidyfastThe initial versions of the nesting and unnesting functions were shown in a preprint. Herein is shown some simple applications and the functions’ speed/efficiency.
library(tidyfast)The following data table will be used for the nesting/unnesting examples.
set.seed(84322)
library(data.table)
library(dplyr) # to compare with case_when()
library(tidyr) # to compare with fill() and separate()
library(ggplot2) # figures
library(ggbeeswarm) # figures
dt <- data.table(
x = rnorm(1e5),
y = runif(1e5),
grp = sample(1L:5L, 1e5, replace = TRUE),
nested1 = lapply(1:10, sample, 10, replace = TRUE),
nested2 = lapply(c("thing1", "thing2"), sample, 10, replace = TRUE),
id = 1:1e5)To make all the comparisons herein more equal, we will set the number
of threads that data.table will use to 1.
setDTthreads(1)We can nest this data using dt_nest():
nested <- dt_nest(dt, grp)
nested
#> Key: <grp>
#> grp data
#> <int> <list>
#> 1: 1 <data.table[19638x5]>
#> 2: 2 <data.table[19987x5]>
#> 3: 3 <data.table[20033x5]>
#> 4: 4 <data.table[20269x5]>
#> 5: 5 <data.table[20073x5]>We can also unnest this with dt_unnest():
dt_unnest(nested, col = data)
#> Key: <grp>
#> grp x y nested1
#> <int> <num> <num> <list>
#> 1: 1 -1.1813164 0.004599736 2,2,1,2,1,1,...
#> 2: 1 -1.0384420 0.853208540 2,8,4,6,7,7,...
#> 3: 1 -0.6247028 0.072652533 4,2,2,1,1,1,...
#> 4: 1 -1.3651514 0.569079215 1,1,1,3,6,2,...
#> 5: 1 0.1403744 0.864617284 10, 1, 1, 1, 8, 1,...
#> ---
#> 99996: 5 -0.3437795 0.995197776 2,1,2,2,2,1,...
#> 99997: 5 1.6157744 0.241735719 10, 1, 1, 1, 8, 1,...
#> 99998: 5 -0.1321246 0.885283934 2,3,3,2,2,4,...
#> 99999: 5 -1.7019715 0.524621296 5,4,3,3,3,2,...
#> 100000: 5 0.3821493 0.032851280 2,8,4,6,7,7,...
#> nested2 id
#> <list> <int>
#> 1: thing2,thing2,thing2,thing2,thing2,thing2,... 2
#> 2: thing2,thing2,thing2,thing2,thing2,thing2,... 8
#> 3: thing1,thing1,thing1,thing1,thing1,thing1,... 15
#> 4: thing1,thing1,thing1,thing1,thing1,thing1,... 17
#> 5: thing2,thing2,thing2,thing2,thing2,thing2,... 20
#> ---
#> 99996: thing1,thing1,thing1,thing1,thing1,thing1,... 99983
#> 99997: thing2,thing2,thing2,thing2,thing2,thing2,... 99990
#> 99998: thing2,thing2,thing2,thing2,thing2,thing2,... 99994
#> 99999: thing2,thing2,thing2,thing2,thing2,thing2,... 99996
#> 100000: thing2,thing2,thing2,thing2,thing2,thing2,... 99998
#> data
#> <list>
#> 1: <data.table[19638x5]>
#> 2: <data.table[19638x5]>
#> 3: <data.table[19638x5]>
#> 4: <data.table[19638x5]>
#> 5: <data.table[19638x5]>
#> ---
#> 99996: <data.table[20073x5]>
#> 99997: <data.table[20073x5]>
#> 99998: <data.table[20073x5]>
#> 99999: <data.table[20073x5]>
#> 100000: <data.table[20073x5]>When our list columns don’t have data tables (as output from
dt_nest()) we can use the dt_hoist() function,
that will unnest vectors. It keeps all the other variables that are not
list-columns as well.
dt_hoist(dt, nested1, nested2)
#> x y grp id nested1 nested2
#> <num> <num> <int> <int> <int> <char>
#> 1: 0.1720703 0.3376675 2 1 1 thing1
#> 2: 0.1720703 0.3376675 2 1 1 thing1
#> 3: 0.1720703 0.3376675 2 1 1 thing1
#> 4: 0.1720703 0.3376675 2 1 1 thing1
#> 5: 0.1720703 0.3376675 2 1 1 thing1
#> ---
#> 999996: 0.6268181 0.7851774 1 100000 1 thing2
#> 999997: 0.6268181 0.7851774 1 100000 5 thing2
#> 999998: 0.6268181 0.7851774 1 100000 7 thing2
#> 999999: 0.6268181 0.7851774 1 100000 6 thing2
#> 1000000: 0.6268181 0.7851774 1 100000 7 thing2Speed comparisons (similar to those shown in the preprint) are
highlighted below. Notably, the timings are without the
nested1 and nested2 columns of the original
dt object from above. Also, all dplyr and
tidyr functions use a tbl version of the
dt table.
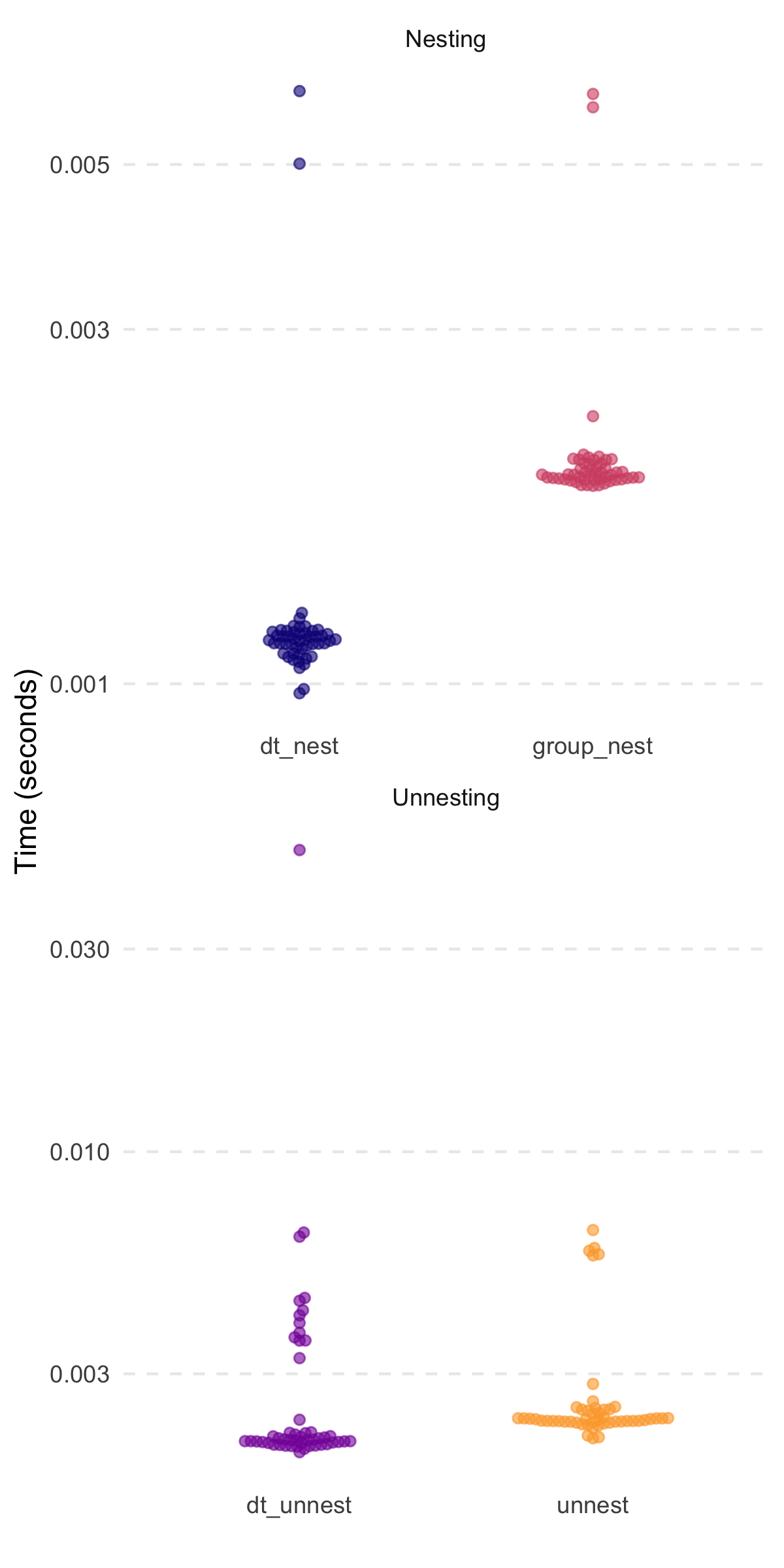
#> # A tibble: 2 × 3
#> expression median mem_alloc
#> <chr> <bch:tm> <bch:byt>
#> 1 dt_nest 1.14ms 2.88MB
#> 2 group_nest 1.91ms 5.12MB
#> # A tibble: 2 × 3
#> expression median mem_alloc
#> <chr> <bch:tm> <bch:byt>
#> 1 dt_unnest 2.08ms 11.84MB
#> 2 unnest 2.33ms 5.96MBThanks to @markfairbanks,
we now have pivoting translations to data.table::melt() and
data.table::dcast(). Consider the following example
(similar to the example in tidyr::pivot_longer() and
tidyr::pivot_wider()):
billboard <- tidyr::billboard
# note the warning - melt is telling us what
# it did with the various data types---logical (where there were just NAs
# and numeric
longer <- billboard %>%
dt_pivot_longer(
cols = c(-artist, -track, -date.entered),
names_to = "week",
values_to = "rank"
)
#> Warning in melt.data.table(data = dt_, id.vars = id_vars, measure.vars = cols,
#> : 'measure.vars' [wk1, wk2, wk3, wk4, ...] are not all of the same type. By
#> order of hierarchy, the molten data value column will be of type 'double'. All
#> measure variables not of type 'double' will be coerced too. Check DETAILS in
#> ?melt.data.table for more on coercion.
longer
#> artist track date.entered week rank
#> <char> <char> <Date> <char> <num>
#> 1: 2 Pac Baby Don't Cry (Keep... 2000-02-26 wk1 87
#> 2: 2Ge+her The Hardest Part Of ... 2000-09-02 wk1 91
#> 3: 3 Doors Down Kryptonite 2000-04-08 wk1 81
#> 4: 3 Doors Down Loser 2000-10-21 wk1 76
#> 5: 504 Boyz Wobble Wobble 2000-04-15 wk1 57
#> ---
#> 24088: Yankee Grey Another Nine Minutes 2000-04-29 wk76 NA
#> 24089: Yearwood, Trisha Real Live Woman 2000-04-01 wk76 NA
#> 24090: Ying Yang Twins Whistle While You Tw... 2000-03-18 wk76 NA
#> 24091: Zombie Nation Kernkraft 400 2000-09-02 wk76 NA
#> 24092: matchbox twenty Bent 2000-04-29 wk76 NA
wider <- longer %>%
dt_pivot_wider(
names_from = week,
values_from = rank
)
wider[, .(artist, track, wk1, wk2)]
#> artist track wk1 wk2
#> <char> <char> <num> <num>
#> 1: 2 Pac Baby Don't Cry (Keep... 87 82
#> 2: 2Ge+her The Hardest Part Of ... 91 87
#> 3: 3 Doors Down Kryptonite 81 70
#> 4: 3 Doors Down Loser 76 76
#> 5: 504 Boyz Wobble Wobble 57 34
#> ---
#> 313: Yankee Grey Another Nine Minutes 86 83
#> 314: Yearwood, Trisha Real Live Woman 85 83
#> 315: Ying Yang Twins Whistle While You Tw... 95 94
#> 316: Zombie Nation Kernkraft 400 99 99
#> 317: matchbox twenty Bent 60 37Notably, there are some current limitations to these: 1)
tidyselect techniques do not work across the board
(e.g. cannot use start_with() and friends) and 2) the
functions are new and likely prone to edge-case bugs.
But let’s compare some basic speed and efficiency. Because of the
data.table functions, these are extremely fast and
efficient.
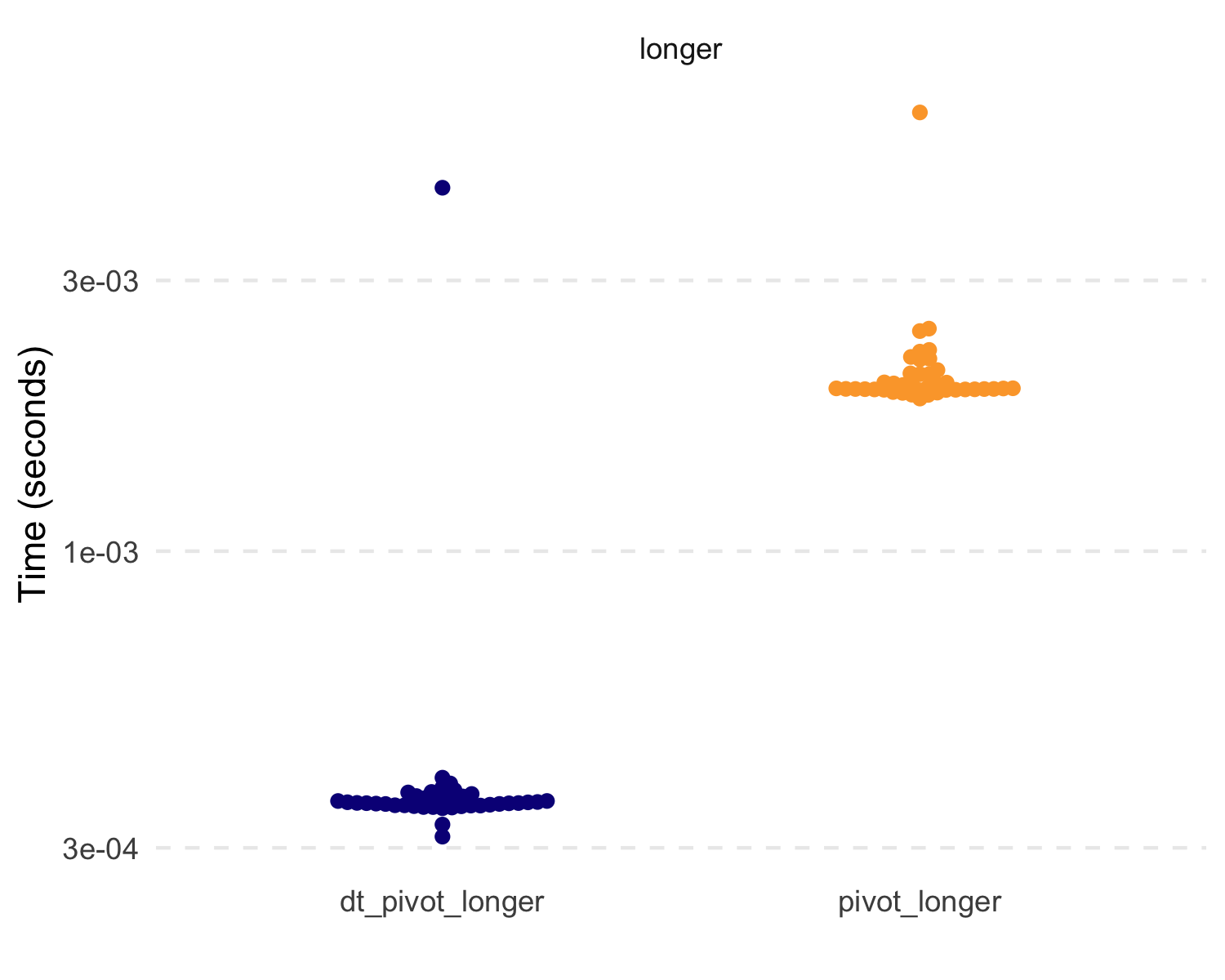
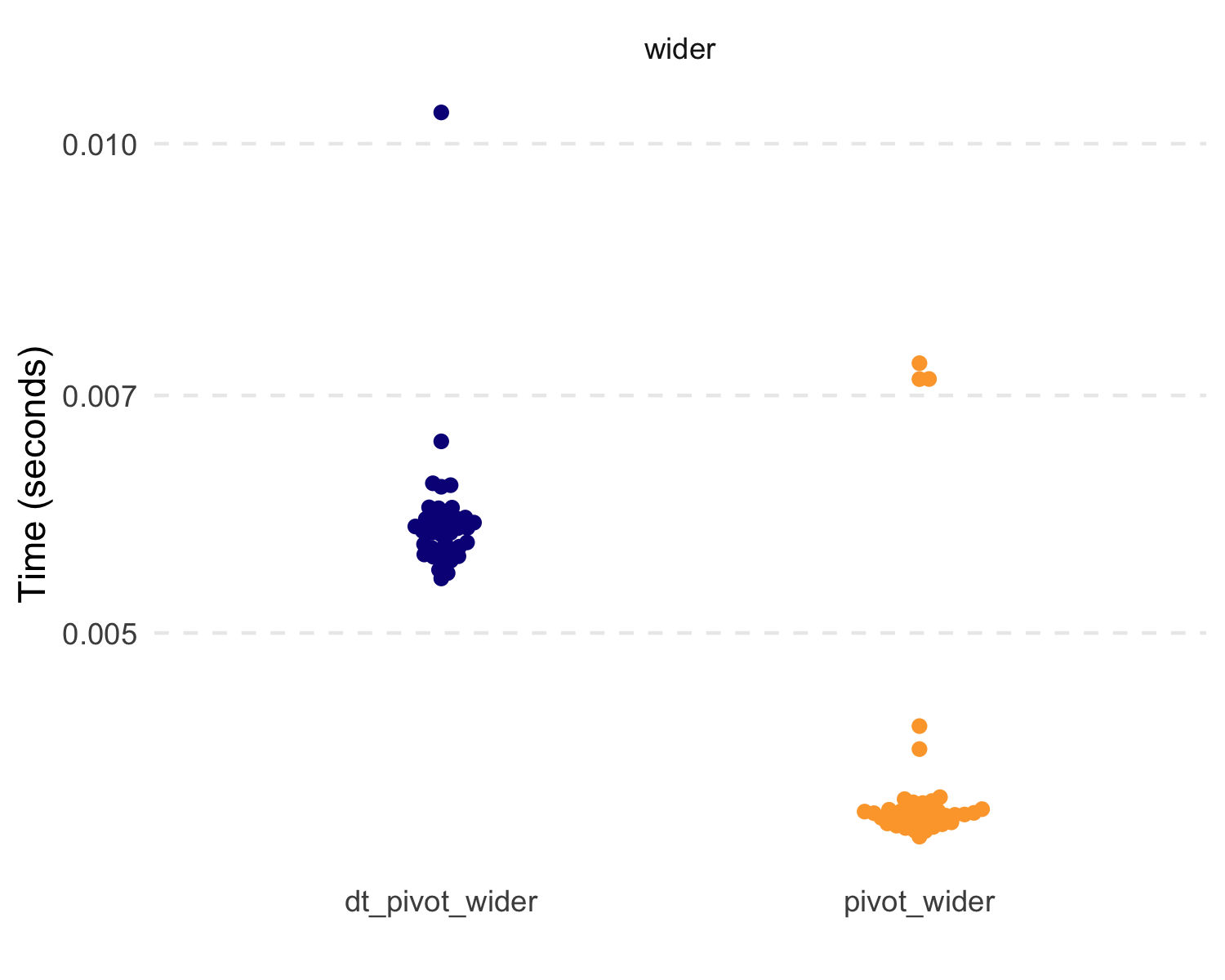
#> # A tibble: 4 × 3
#> expression median mem_alloc
#> <chr> <bch:tm> <bch:byt>
#> 1 dt_pivot_longer 359.98µs 1001.23KB
#> 2 pivot_longer 1.94ms 1.73MB
#> 3 dt_pivot_wider 5.8ms 1.99MB
#> 4 pivot_wider 3.87ms 2.71MBAlso, the new dt_case_when() function is built on the
very fast data.table::fiflese() but has syntax like unto
dplyr::case_when(). That is, it looks like:
dt_case_when(condition1 ~ label1,
condition2 ~ label2,
...)To show that each method, dt_case_when(),
dplyr::case_when(), and data.table::fifelse()
produce the same result, consider the following example.
x <- rnorm(1e6)
medianx <- median(x)
x_cat <-
dt_case_when(x < medianx ~ "low",
x >= medianx ~ "high",
is.na(x) ~ "unknown")
x_cat_dplyr <-
case_when(x < medianx ~ "low",
x >= medianx ~ "high",
is.na(x) ~ "unknown")
x_cat_fif <-
fifelse(x < medianx, "low",
fifelse(x >= medianx, "high",
fifelse(is.na(x), "unknown", NA_character_)))
identical(x_cat, x_cat_dplyr)
#> [1] TRUE
identical(x_cat, x_cat_fif)
#> [1] TRUENotably, dt_case_when() is very fast and memory
efficient, given it is built on data.table::fifelse().
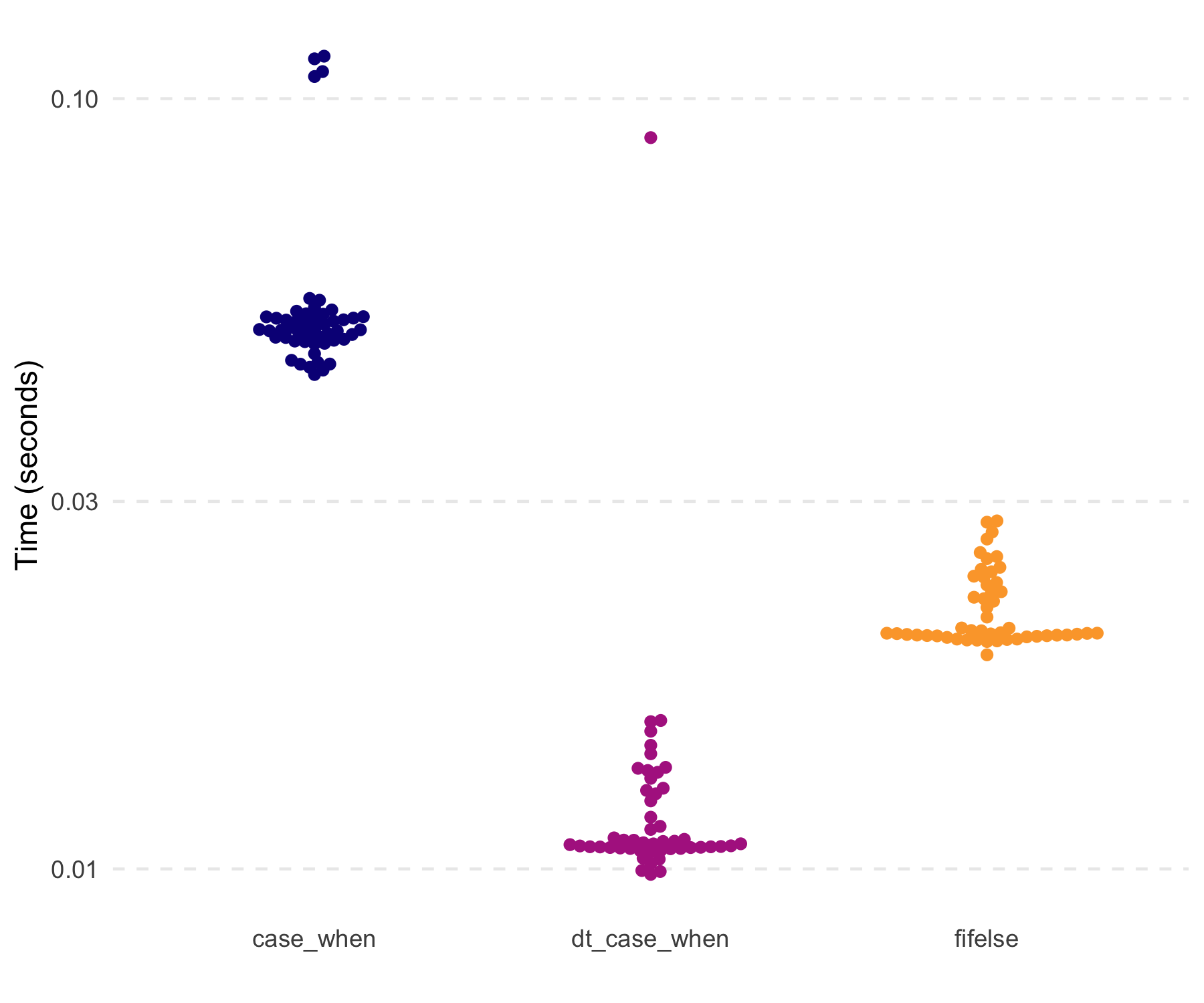
#> # A tibble: 3 × 3
#> expression median mem_alloc
#> <chr> <bch:tm> <bch:byt>
#> 1 case_when 45.2ms 72.5MB
#> 2 dt_case_when 10.7ms 19.1MB
#> 3 fifelse 20.1ms 34.3MBA new function is dt_fill(), which fulfills the role of
tidyr::fill() to fill in NA values with values
around it (either the value above, below, or trying both). This
currently relies on the efficient C++ code from
tidyr (fillUp() and
fillDown()).
x = 1:10
dt_with_nas <- data.table(
x = x,
y = shift(x, 2L),
z = shift(x, -2L),
a = sample(c(rep(NA, 10), x), 10),
id = sample(1:3, 10, replace = TRUE)
)
# Original
dt_with_nas
#> x y z a id
#> <int> <int> <int> <int> <int>
#> 1: 1 NA 3 NA 3
#> 2: 2 NA 4 9 3
#> 3: 3 1 5 NA 1
#> 4: 4 2 6 8 3
#> 5: 5 3 7 NA 2
#> 6: 6 4 8 NA 2
#> 7: 7 5 9 7 3
#> 8: 8 6 10 NA 2
#> 9: 9 7 NA NA 2
#> 10: 10 8 NA 4 2
# All defaults
dt_fill(dt_with_nas, y, z, a, immutable = FALSE)
#> x y z a id
#> <int> <int> <int> <int> <int>
#> 1: 1 NA 3 NA 3
#> 2: 2 NA 4 9 3
#> 3: 3 1 5 9 1
#> 4: 4 2 6 8 3
#> 5: 5 3 7 8 2
#> 6: 6 4 8 8 2
#> 7: 7 5 9 7 3
#> 8: 8 6 10 7 2
#> 9: 9 7 10 7 2
#> 10: 10 8 10 4 2
# by id variable called `grp`
dt_fill(dt_with_nas,
y, z, a,
id = list(id))
#> x y z a id
#> <int> <int> <int> <int> <int>
#> 1: 1 NA 3 NA 3
#> 2: 2 NA 4 9 3
#> 3: 3 1 5 9 1
#> 4: 4 2 6 8 3
#> 5: 5 3 7 8 2
#> 6: 6 4 8 8 2
#> 7: 7 5 9 7 3
#> 8: 8 6 10 7 2
#> 9: 9 7 10 7 2
#> 10: 10 8 10 4 2
# both down and then up filling by group
dt_fill(dt_with_nas,
y, z, a,
id = list(id),
.direction = "downup")
#> x y z a id
#> <int> <int> <int> <int> <int>
#> 1: 1 2 3 9 3
#> 2: 2 2 4 9 3
#> 3: 3 1 5 9 1
#> 4: 4 2 6 8 3
#> 5: 5 3 7 8 2
#> 6: 6 4 8 8 2
#> 7: 7 5 9 7 3
#> 8: 8 6 10 7 2
#> 9: 9 7 10 7 2
#> 10: 10 8 10 4 2In its current form, dt_fill() is faster than
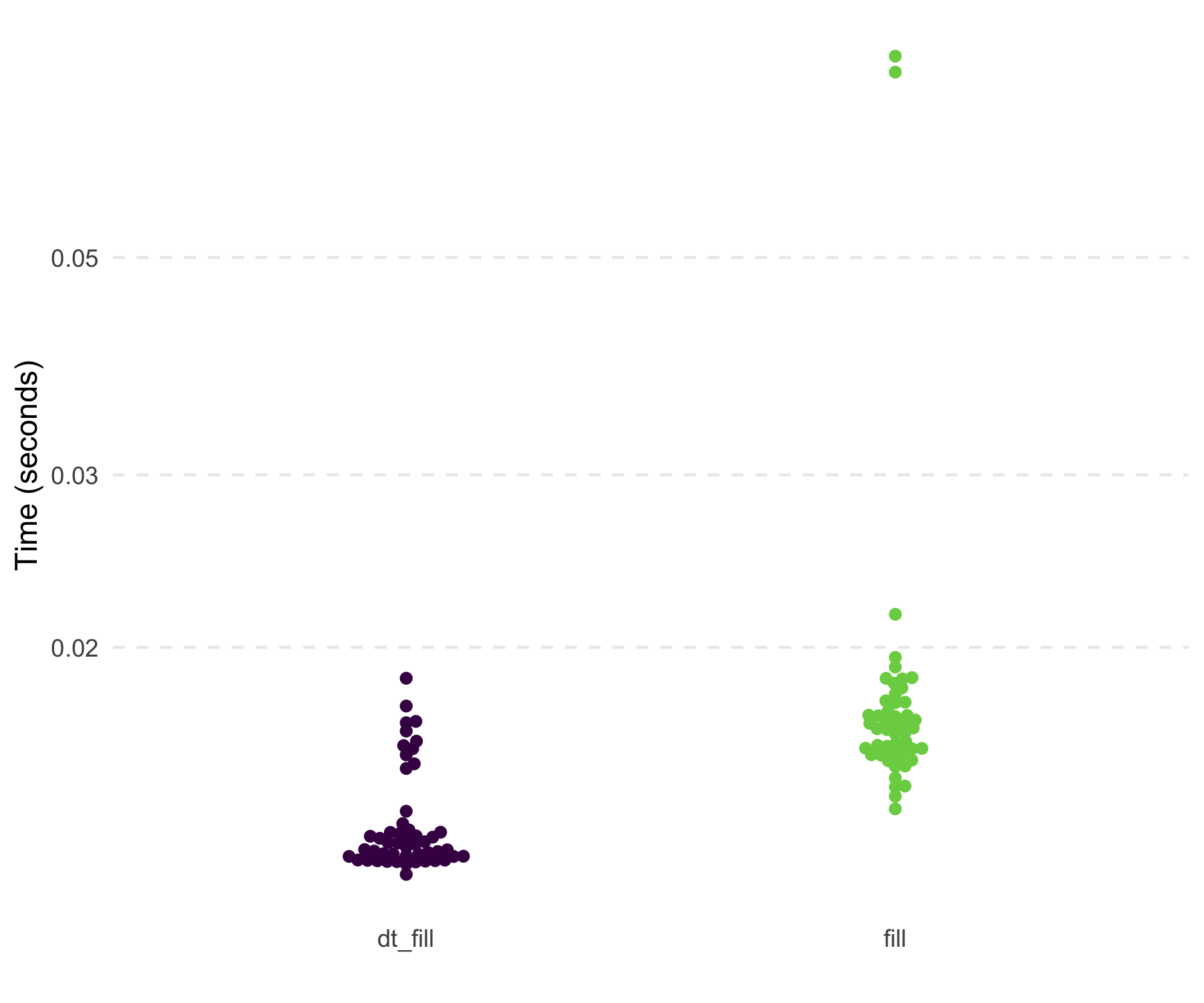
tidyr::fill() and uses slightly less memory. Below are the
results of filling in the NAs within each id
on a 19 MB data set.
x = 1:1e6
dt3 <- data.table(
x = x,
y = shift(x, 10L),
z = shift(x, -10L),
a = sample(c(rep(NA, 10), x), 10),
id = sample(1:3, 10, replace = TRUE))
df3 <- data.frame(dt3)
marks3 <-
bench::mark(
tidyr::fill(dplyr::group_by(df3, id), x, y),
tidyfast::dt_fill(dt3, x, y, id = list(id)),
check = FALSE,
iterations = 50
)
#> # A tibble: 2 × 3
#> expression median mem_alloc
#> <bch:expr> <bch:tm> <bch:byt>
#> 1 tidyr::fill(dplyr::group_by(df3, id), x, y) 15.4ms 46.4MB
#> 2 tidyfast::dt_fill(dt3, x, y, id = list(id)) 12.4ms 17.6MBThe dt_separate() function is still under heavy
development. Its behavior is similar to tidyr::separate()
but is lacking some functionality currently. For example,
into needs to be supplied the maximum number of possible
columns to separate.
dt_separate(data.table(col = "A.B.C"), col, into = c("A", "B"))
#> Error in `[.data.table`(dt, , eval(split_it)) :
#> Supplied 2 columns to be assigned 3 items. Please see NEWS for v1.12.2.For current functionality, consider the following example.
dt_to_split <- data.table(
x = paste(letters, LETTERS, sep = ".")
)
dt_separate(dt_to_split, x, into = c("lower", "upper"))#> lower upper
#> <char> <char>
#> 1: a A
#> 2: b B
#> 3: c C
#> 4: d D
#> 5: e E
#> 6: f FTesting with a 4 MB data set with one variable that has columns of
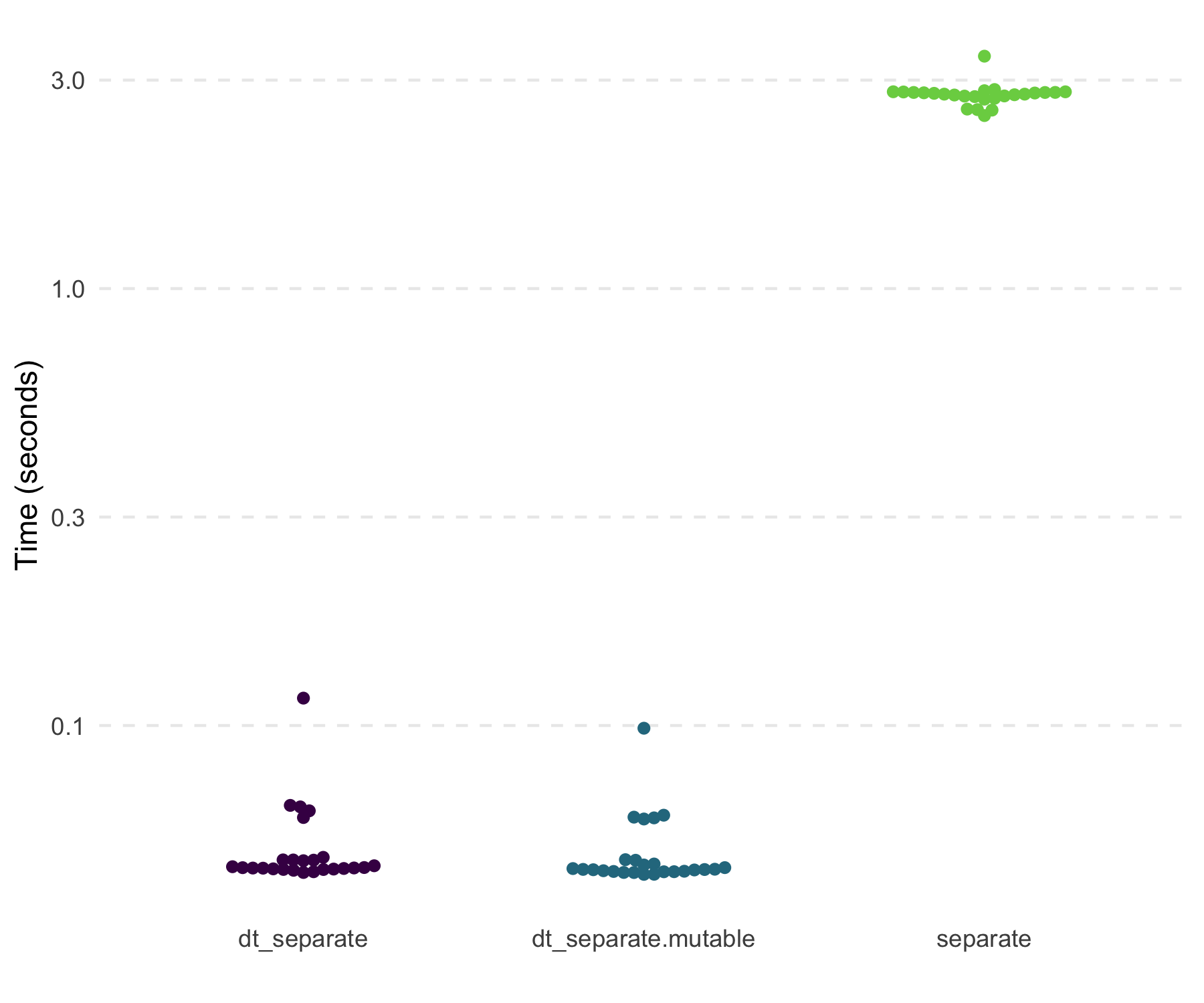
“A.B” repeatedly, shows that dt_separate() is fast and far
more memory efficient compared to tidyr::separate().
#> # A tibble: 3 × 3
#> expression median mem_alloc
#> <chr> <bch:tm> <bch:byt>
#> 1 separate 2.79s 3.89GB
#> 2 dt_separate 47.31ms 26.73MB
#> 3 dt_separate-mutable 46.85ms 26.72MBThe dt_count() function does essentially what
dplyr::count() does. Notably, this, unlike the majority of
other dt_ functions, wraps a very simple statement in
data.table. That is, data.table makes getting
counts very simple and concise. Nonetheless, dt_count()
fits the general API of tidyfast. To some degree,
dt_uncount() is also a fairly simple wrapper, although the
approach may not be as straightforward as that for
dt_count().
The following examples show how count and uncount can work. We’ll use
the dt data table from the nesting examples.
counted <- dt_count(dt, grp)
counted
#> Key: <grp>
#> grp N
#> <int> <int>
#> 1: 1 19638
#> 2: 2 19987
#> 3: 3 20033
#> 4: 4 20269
#> 5: 5 20073uncounted <- dt_uncount(counted, N)
uncounted[]
#> Key: <grp>
#> grp
#> <int>
#> 1: 1
#> 2: 1
#> 3: 1
#> 4: 1
#> 5: 1
#> ---
#> 99996: 5
#> 99997: 5
#> 99998: 5
#> 99999: 5
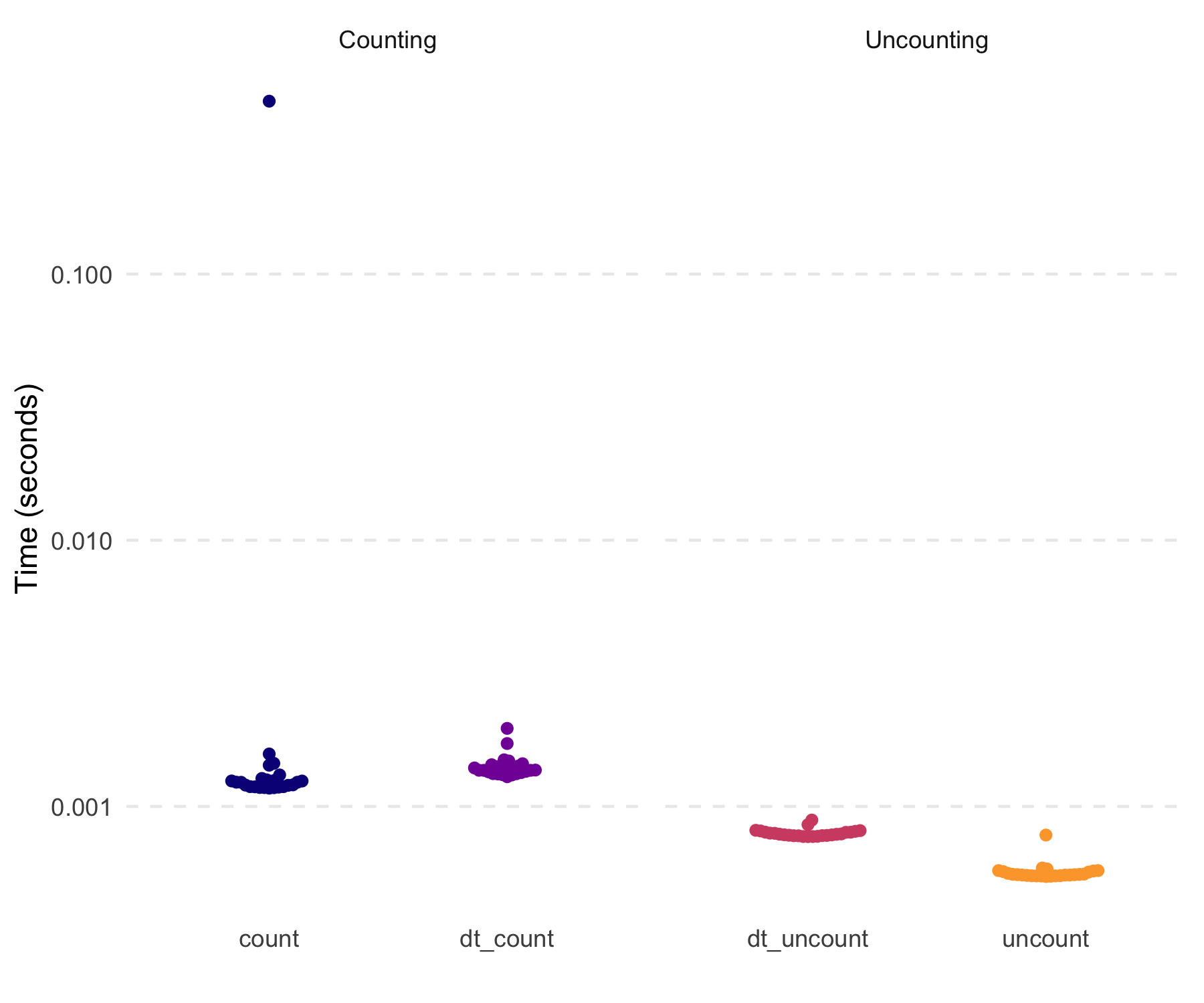
#> 100000: 5These are also quick (not that the tidyverse functions
were at all slow here).
dt5 <- copy(dt)
df5 <- data.frame(dt5)
marks5 <-
bench::mark(
counted_tbl <- dplyr::count(df5, grp),
counted_dt <- tidyfast::dt_count(dt5, grp),
tidyr::uncount(counted_tbl, n),
tidyfast::dt_uncount(counted_dt, N),
check = FALSE,
iterations = 25
)
Please note that the tidyfast project is released with a
Contributor
Code of Conduct. By contributing to this project, you agree to abide
by its terms.
We want to thank our wonderful contributors:
tidytable
package that compliments some of tidyfasts
functionality.Complementary Packages:
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.