
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The tidycensuskr package is designed for R users who
want to work with South Korean census and administrative boundary data.
It aims to provide an easy-to-use interface for population, housing, and
socioeconomic statistics linked with geospatial boundaries.
You can install the released version of tidycensuskr
from CRAN with:
# CRAN
install.packages("tidycensuskr")
# R-universe
install.packages("tidycensuskr", repos = "https://sigmafelix.r-universe.dev")To install the development version,
remotes::install_github() will suffice.
# Development version from GitHub
rlang::check_installed("remotes")
remotes::install_github("sigmafelix/tidycensuskr")As of September 2025, this package contains two datasets: Census data
(censuskor) and the corresponding geospatial data.
anycensus()anycensus() allows you to query census
data for specific district or province codes and types of data
(population, tax, mortality, economy, housing) for three census years
(2010, 2015, 2020).# loading Seoul population data
tidycensuskr::anycensus(codes = "Seoul", type = "population")
#> # A tibble: 25 × 17
#> year adm1 adm1_code adm2 adm2_code type all households_total…¹
#> <dbl> <chr> <dbl> <chr> <dbl> <chr> <dbl>
#> 1 2020 Seoul 11 Dobong-gu 11100 populat… 312878
#> 2 2020 Seoul 11 Dongdaemun-gu 11060 populat… 332796
#> 3 2020 Seoul 11 Dongjak-gu 11200 populat… 378749
#> 4 2020 Seoul 11 Eunpyeong-gu 11120 populat… 458777
#> 5 2020 Seoul 11 Gangbuk-gu 11090 populat… 295304
#> 6 2020 Seoul 11 Gangdong-gu 11250 populat… 440022
#> 7 2020 Seoul 11 Gangnam-gu 11230 populat… 509899
#> 8 2020 Seoul 11 Gangseo-gu 11160 populat… 564114
#> 9 2020 Seoul 11 Geumcheon-gu 11180 populat… 225594
#> 10 2020 Seoul 11 Guro-gu 11170 populat… 394733
#> # ℹ 15 more rows
#> # ℹ abbreviated name: ¹`all households_total_prs`
#> # ℹ 10 more variables: `all households_male_prs` <dbl>,
#> # `all households_female_prs` <dbl>, fertility_total_brt <dbl>,
#> # `fertility_15-19 (simulated)_bp1` <dbl>, `fertility_20-24_bp1` <dbl>,
#> # `fertility_25-29_bp1` <dbl>, `fertility_30-34_bp1` <dbl>,
#> # `fertility_35-39_bp1` <dbl>, `fertility_40-44_bp1` <dbl>, …censuskordata(censuskor) loads an attached dataset
that contains the census data in long form. This dataset is
automatically loaded upon loading the package.load_district()load_district() allows you to get the
Si-Gun-Gu level sf files for the three census
years (2010, 2015, 2020).tidycensuskr.sf package to be
installed. Please install it from R-universe using
install.packages("tidycensuskr.sf", repos = "https://sigmafelix.r-universe.dev").# loading boundary sf file: 2020 boundaries are included in this package
data(adm2_sf_2020)
# tidycensuskr.sf::load_districts(year = 2020)Package vignettes are the first place to look for detailed examples. Below are some quick examples to get you started.
anycensus() will return an analysis-ready data.frame
that can be easily merged with the corresponding boundary
sf object from load_districts(). Here is a
simple example of making maps with population data.
library(tidycensuskr)
#> tidycensuskr 0.2.6 (2025-12-09)
#> Please install the companion data package tidycensuskr.sf to use the district boundaries.
#> install.packages('tidycensuskr.sf', repos = 'https://sigmafelix.r-universe.dev')
library(ggplot2)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(tidyr)
library(sf)
#> Linking to GEOS 3.12.2, GDAL 3.11.4, PROJ 9.4.1; sf_use_s2() is TRUE
library(biscale)
library(cowplot)
sf_use_s2(FALSE)
#> Spherical geometry (s2) switched off
options(scipen = 100)
# load census data
census_pop_2020 <- anycensus(year = 2020, codes = NULL, type = "population")
#> Using character codes that are convertible to integers. Automatically converting to integers...
census_pop_2020 <- census_pop_2020 |>
rename(population_total = `all households_total_prs`)
# load boundaries
data(adm2_sf_2020)
adm2_2020 <- adm2_sf_2020
# merge boundaries and census data
census_2020_sf <- adm2_2020 |>
left_join(census_pop_2020, by = c("adm2_code" = "adm2_code"))
# plot population data
census_2020_pop <-
ggplot(census_2020_sf) +
geom_sf(aes(fill = population_total), color = "white", size = 0.1) +
theme_minimal() +
labs(
title = "Population (2020)",
fill = "Population"
) +
theme(
plot.title = element_text(size = 12),
axis.text = element_text(size = 7),
legend.text = element_text(size = 7),
legend.title = element_text(size = 8)
)
census_2020_pop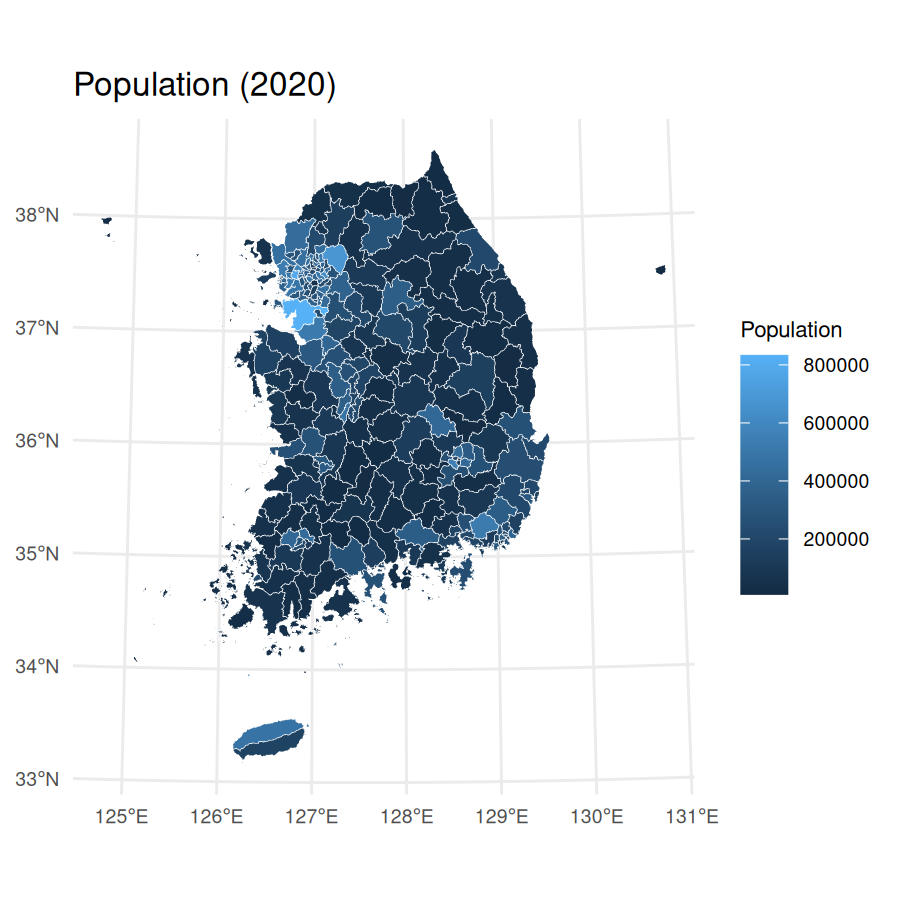
For Seoul Metropolitan Area (including Seoul, Incheon, and
Gyeonggi-do), you can use a character vector in codes
argument and merge the retrieved data.frame and
sf object with inner_join():
census_pop_2020_sma <-
anycensus(
year = 2020,
codes = c("Seoul", "Incheon", "Gyeonggi"),
type = "population"
) |>
rename(population_total = `all households_total_prs`)
census_2020_sf_sma <- adm2_2020 |>
inner_join(census_pop_2020_sma, by = c("year", "adm2_code"))
# plot population data
census_2020_pop_sma <-
ggplot(census_2020_sf_sma) +
geom_sf(aes(fill = population_total), color = "white", size = 0.1) +
theme_minimal() +
labs(
title = "Population in Seoul Metropolitan Area (2020)",
fill = "Population"
) +
theme(
plot.title = element_text(size = 12),
axis.text = element_text(size = 7),
legend.text = element_text(size = 7),
legend.title = element_text(size = 8)
)
census_2020_pop_sma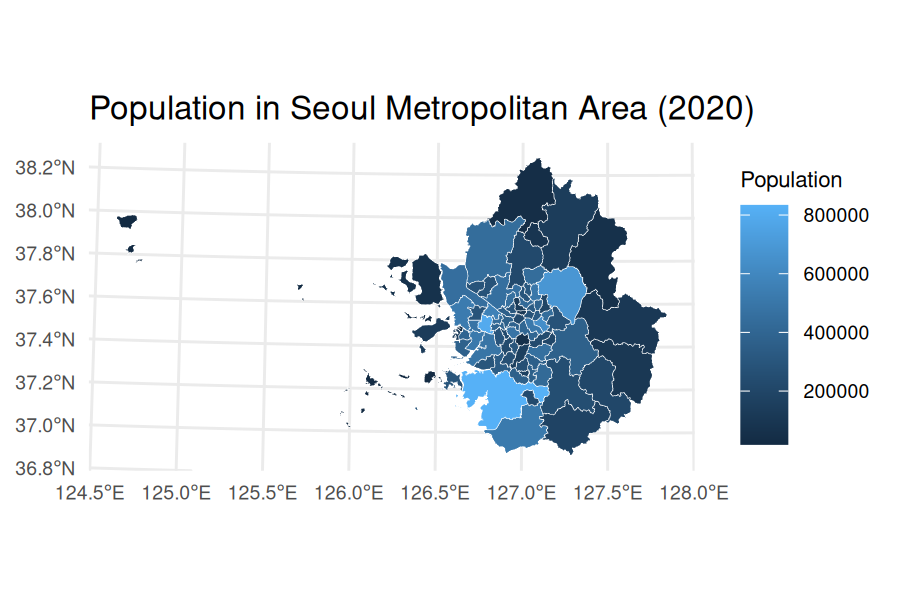
Moving on to a complex example, the code below demonstrates to generate a bivariate map with persons per housing unit and all-cause mortality rate.
census_housing_2020 <- anycensus(year = 2020, codes = NULL, type = "housing")
#> Using character codes that are convertible to integers. Automatically converting to integers...
census_housing_2020 <- census_housing_2020 |>
rename(housing_total_units = `housing types_total_cnt`)
census_pop_housing_2020 <- census_pop_2020 |>
left_join(census_housing_2020 |>
select(adm2_code, housing_total_units),
by = "adm2_code") |>
transmute(
adm2_code = adm2_code,
persons_per_housing = population_total / housing_total_units
)
census_mort_2020 <- anycensus(year = 2020, codes = NULL, type = "mortality")
#> Using character codes that are convertible to integers. Automatically converting to integers...
census_mort_2020 <- census_mort_2020 |>
rename(mortality_total = `all causes_total_p1p`)
census_pph_mort_2020 <- census_pop_housing_2020 |>
left_join(census_mort_2020 |>
select(adm2_code, mortality_total),
by = "adm2_code")
# merge boundaries and census data
census_2020_sf <- adm2_2020 |>
left_join(census_pph_mort_2020, by = c("adm2_code" = "adm2_code"))
census_2020_mapbase <-
biscale::bi_class(
census_2020_sf,
x = persons_per_housing,
y = mortality_total,
style = "quantile",
dim = 3
)
# draw a bivariate legend
legend <- bi_legend(pal = "DkCyan",
dim = 3,
xlab = "More Persons per Housing ",
ylab = "All-Cause Mortality ",
size = 6)
# plot population data
census_2020_bmap <-
ggplot(census_2020_mapbase) +
geom_sf(
aes(fill = bi_class),
color = "white",
size = 0.1,
show.legend = FALSE
) +
bi_scale_fill(pal = "DkCyan", dim = 3) +
theme_minimal() +
labs(title = "Persons per housing unit and all-cause mortality rate (2020)") +
bi_theme(base_size = 10) +
theme(plot.title = element_text(size = 10))
# combine map with legend
census_2020_bimap <- cowplot::ggdraw() +
cowplot::draw_plot(census_2020_bmap, 0, 0, 1, 1) +
cowplot::draw_plot(legend, 0.7, 0.02, 0.3, 0.3)
census_2020_bimap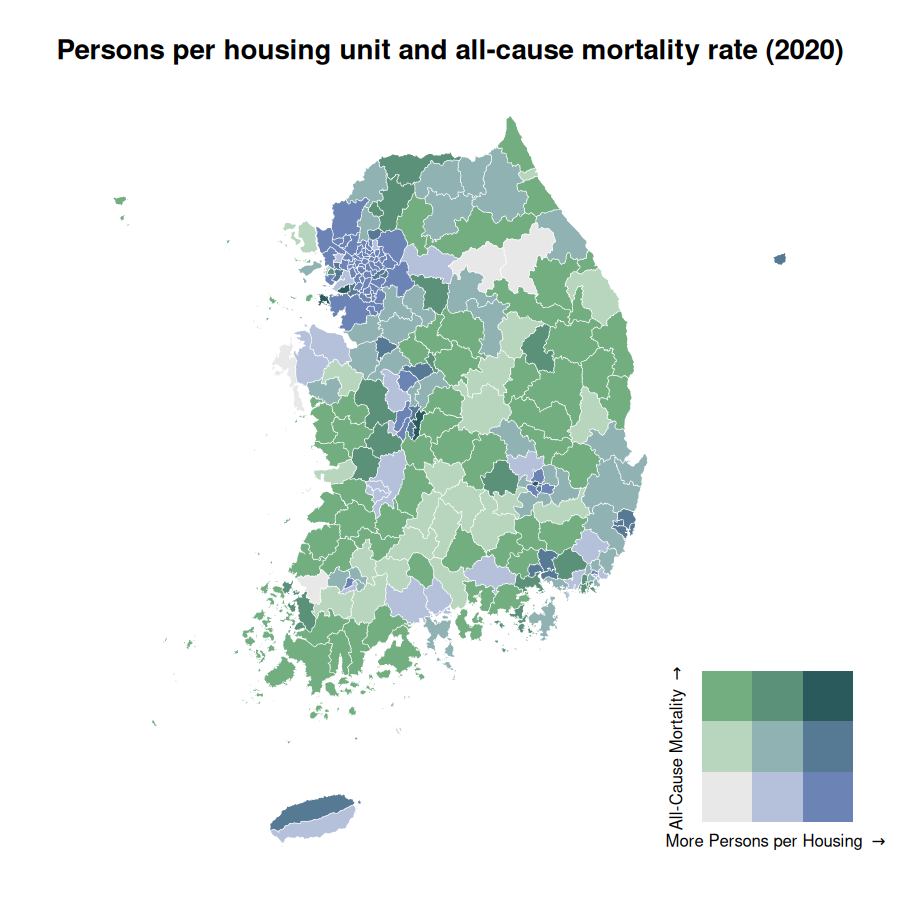
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.