The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Visualize and tabulate single-choice, multiple-choice, matrix-style questions from survey data. Includes ability to group cross-tabulations, frequency distributions, and plots by categorical variables and to integrate survey weights. Ideal for quickly uncovering descriptive patterns in survey data.
install.packages("surveyexplorer")
# or devtools::install_github("liamhaller/surveyexplorer") for the devlopment version library(surveyexplorer)The data used in the following examples is from the
berlinbears dataset, a fictional survey of bears in Berlin,
that is included in the surveyexplorer package.
#Basic table
single_table(berlinbears,
question = income)| Question: income | ||
| <1000 | ||
|---|---|---|
| 1000-2000 | ||
| 2000-3000 | ||
| 3000-4000 | ||
| 5000+ | ||
| No answer | ||
| NA | ||
| Column Total | ||
Use group_by = to partition the question into several
groups
single_table(berlinbears,
question = income,
group_by = gender)| Question: income | ||||||||
| grouped by: gender | ||||||||
| <1000 | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1000-2000 | ||||||||
| 2000-3000 | ||||||||
| 3000-4000 | ||||||||
| 5000+ | ||||||||
| No answer | ||||||||
| NA | ||||||||
| Columnwise Total | ||||||||
Ignore unwanted subgroups with subgroups_to_exclude
single_table(berlinbears,
question = income,
group_by = gender,
subgroups_to_exclude = NA) | Question: income | ||||||
| grouped by: gender | ||||||
| <1000 | ||||||
|---|---|---|---|---|---|---|
| 1000-2000 | ||||||
| 2000-3000 | ||||||
| 3000-4000 | ||||||
| 5000+ | ||||||
| No answer | ||||||
| NA | ||||||
| Columnwise Total | ||||||
Remove NAs from the question variable with na.rm
single_table(berlinbears,
question = income,
group_by = gender,
subgroups_to_exclude = NA,
na.rm = TRUE)| Question: income | ||||||
| grouped by: gender | ||||||
| <1000 | ||||||
|---|---|---|---|---|---|---|
| 1000-2000 | ||||||
| 2000-3000 | ||||||
| 3000-4000 | ||||||
| 5000+ | ||||||
| No answer | ||||||
| Columnwise Total | ||||||
Finally, you can specify survey weights using the weight option
single_table(berlinbears,
question = income,
group_by = gender,
subgroups_to_exclude = NA,
na.rm = TRUE,
weights = weights)| Question: income | ||||||
| grouped by: gender | ||||||
| <1000 | ||||||
|---|---|---|---|---|---|---|
| 1000-2000 | ||||||
| 2000-3000 | ||||||
| 3000-4000 | ||||||
| 5000+ | ||||||
| No answer | ||||||
| Columnwise Total | ||||||
| Frequencies and counts are weighted | ||||||
The same syntax can be applied to the single_freq
function to plot frequencies of the question optionally partitioned by
subgroups.
single_freq(berlinbears,
question = income,
group_by = gender,
subgroups_to_exclude = NA,
na.rm = TRUE,
weights = weights)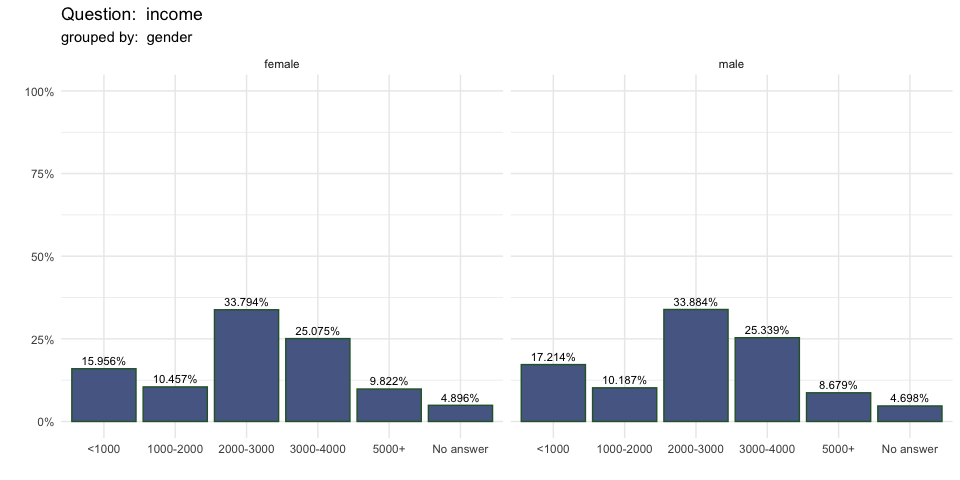
The options and syntax for multiple-choice tables
multi_table and graphs multi_graphs are the
same. The only difference is the question input also accommodates
tidyselect syntax to select several columns for each answer option. For
example, the question “will_eat” has five answer options each prefixed
by “will_eat”
berlinbears |>
dplyr::select(starts_with('will_eat')) |>
head()
#> will_eat.SQ001 will_eat.SQ002 will_eat.SQ003 will_eat.SQ004 will_eat.SQ005
#> 1 0 1 0 1 1
#> 2 0 1 1 1 1
#> 3 1 1 0 1 1
#> 4 0 0 0 1 0
#> 5 0 0 0 1 1
#> 6 0 0 0 1 0The same syntax can be used to select the question for the multiple choice tables and graphs
multi_table(berlinbears,
question = dplyr::starts_with('will_eat'),
group_by = genus,
subgroups_to_exclude = NA,
na.rm = TRUE)| Question: dplyr::starts_with(“will_eat”) | ||||||
| grouped by: genus | ||||||
| will_eat.SQ004 | ||||||
|---|---|---|---|---|---|---|
| will_eat.SQ002 | ||||||
| will_eat.SQ005 | ||||||
| will_eat.SQ001 | ||||||
| will_eat.SQ003 | ||||||
| Columnwise Total | ||||||
For graphing, the multi_freq function creates an UpSet
plot to visualize the frequencies of the intersecting sets for each
answer combination and also includes the ability to specify weights.
multi_freq(berlinbears,
question = dplyr::starts_with('will_eat'),
na.rm = TRUE,
weights = weights)
#> Estimes are only preciese to one significant digit, weights may have been rounded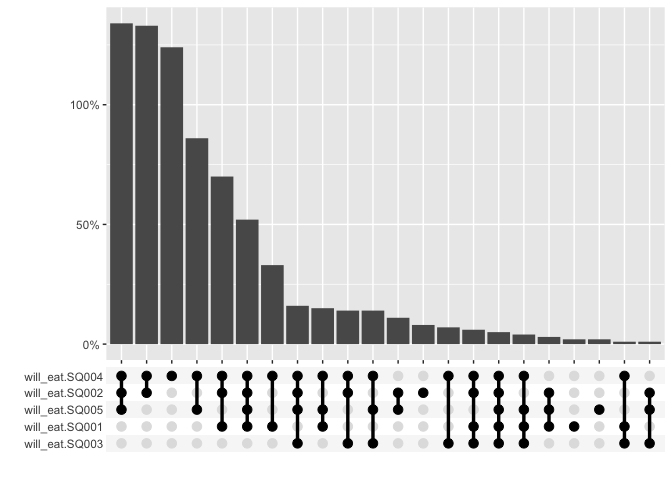
The graphs can also be grouped
multi_freq(berlinbears,
question = dplyr::starts_with('will_eat'),
group_by = genus,
subgroups_to_exclude = NA,
na.rm = FALSE,
weights = weights)
#> Estimes are only preciese to one significant digit, weights may have been rounded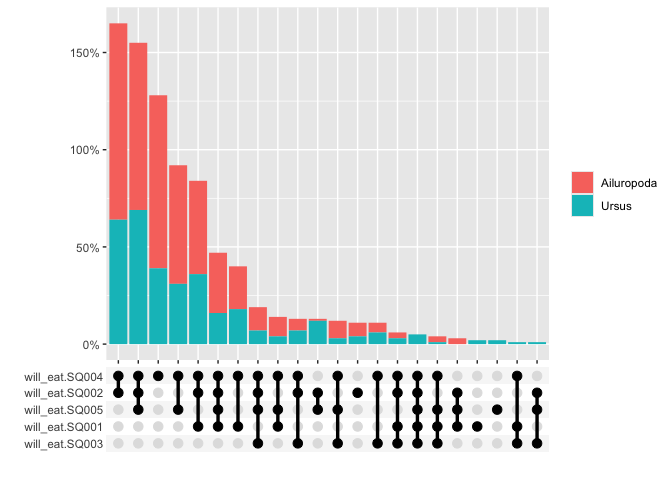
matrix_table has the same syntax as above and works with
array or categorical questions
matrix_table(berlinbears,
dplyr::starts_with('c_'),
group_by = is_parent)| Question: dplyr::starts_with(“c_”) | ||||
| grouped by: is_parent | ||||
| 0 | ||||
|---|---|---|---|---|
| c_diet | ||||
| c_exercise | ||||
| 1 | ||||
| c_diet | ||||
| c_exercise | ||||
matrix_freq visualizes the frequencies of responses
matrix_freq(berlinbears,
dplyr::starts_with('p_'),
na.rm = TRUE)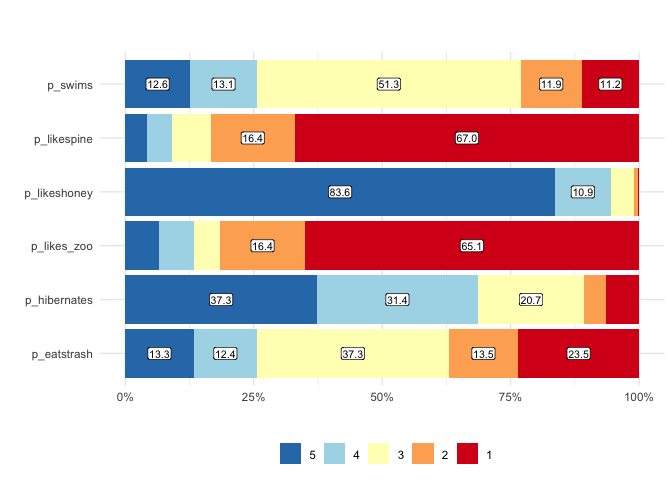
For array/matrix style questions that are numeric
matrix_mean plots the mean values and confidence
intervals
matrix_mean(berlinbears,
question = dplyr::starts_with('p_'),
na.rm = TRUE)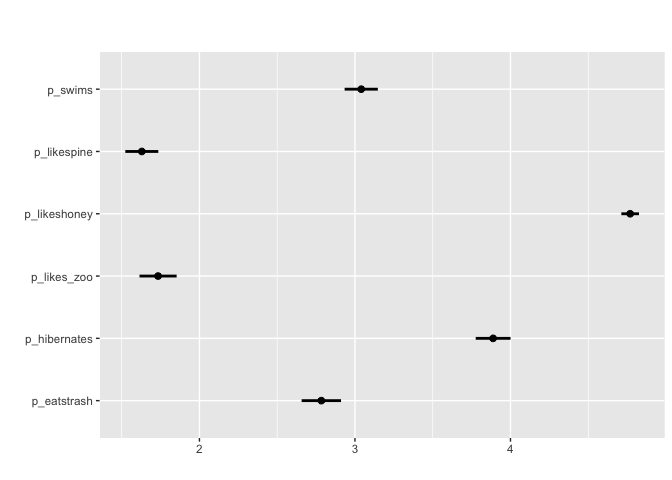
#Can also apply grouping + survey weights
matrix_mean(berlinbears,
question = dplyr::starts_with('p_'),
na.rm = TRUE,
group_by = species,
subgroups_to_exclude = NA)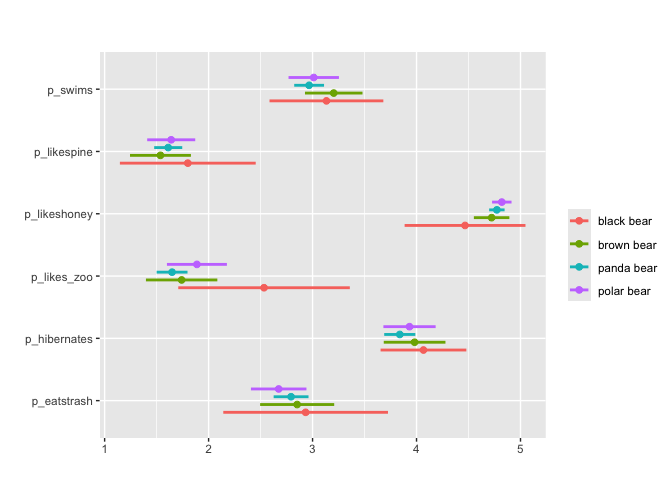
Finally, for Likert questions (scales of 3,5,7,9…)
matrix_likert provides a custom plot
#you can specify custom labels with the `label` argument
matrix_likert(berlinbears,
question = dplyr::starts_with('p_'),
labels = c('Strongly disagree', 'Disagree','Neutral','Agree','Strongly agree'))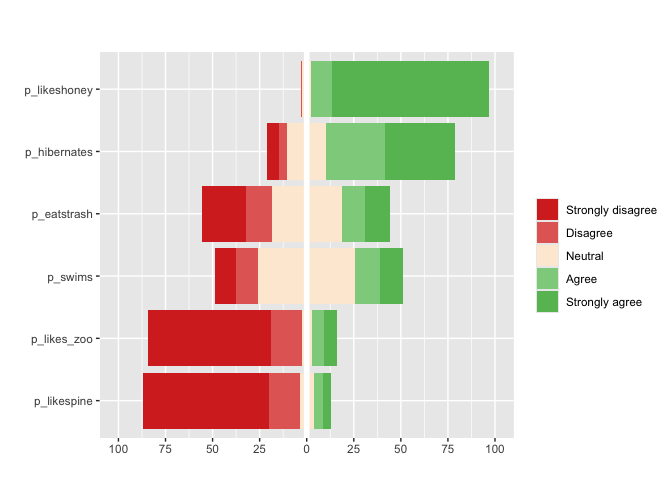
#can also apply pass custom colors and specify weights weights
matrix_likert(berlinbears,
question = dplyr::starts_with('p_'),
labels = c('Strongly disagree', 'Disagree','Neutral','Agree','Strongly agree'),
colors = c("#E1AA28", "#1E5F46", "#7E8F75", "#EFCD83", "#E17832"),
weights = weights) 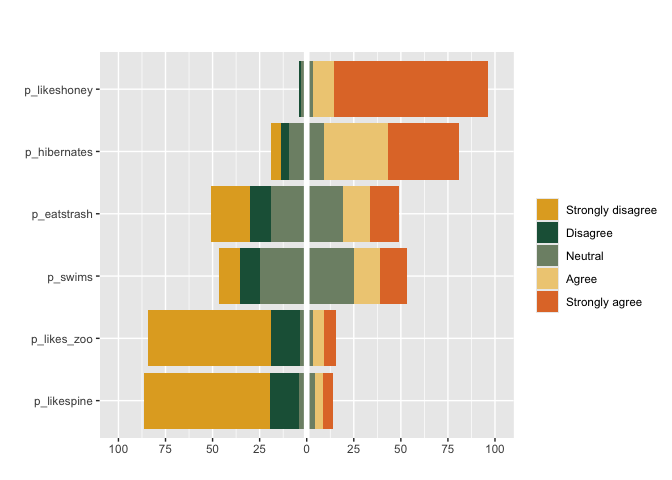
single_tablesingle_freqmulti_tablemulti_freqmatrix_tablematrix_freqmatrix_meanmatrix_likert*_table functions return a gt table of the cross tabulations and frequencies for each question while *_freq returns the same data but as a plot.
For matrix-style questions with numerical input,
matrix_mean plots the mean value value and ± two standard
deviations. matrix_likert visualizes questions that accept
Likert responses (strongly agree-strongly disagree) or questions with
3,5,7,9… categories.
Each function contains the following options
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.