
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

The goal of spatialsample is to provide functions and classes for spatial resampling to use with rsample, including:
Like rsample, spatialsample provides building blocks for creating and analyzing resamples of a spatial data set but does not include code for modeling or computing statistics. The resampled data sets created by spatialsample are efficient and do not have much memory overhead.
You can install the released version of spatialsample from CRAN with:
install.packages("spatialsample")And the development version from GitHub with:
# install.packages("pak")
pak::pak("tidymodels/spatialsample")The most straightforward spatial resampling strategy is
spatial_clustering_cv(), which uses k-means clustering to
identify cross-validation folds:
library(spatialsample)
set.seed(1234)
folds <- spatial_clustering_cv(boston_canopy, v = 5)
folds
#> # 5-fold spatial cross-validation
#> # A tibble: 5 × 2
#> splits id
#> <list> <chr>
#> 1 <split [604/78]> Fold1
#> 2 <split [595/87]> Fold2
#> 3 <split [524/158]> Fold3
#> 4 <split [490/192]> Fold4
#> 5 <split [515/167]> Fold5In this example, the boston_canopy data on tree cover in
Boston, MA is resampled with v = 5; notice that the
resulting partitions do not contain an equal number of observations.
In addition to resampling algorithms, spatialsample provides methods
to visualize resamples using ggplot2 through the
autoplot() function:
autoplot(folds)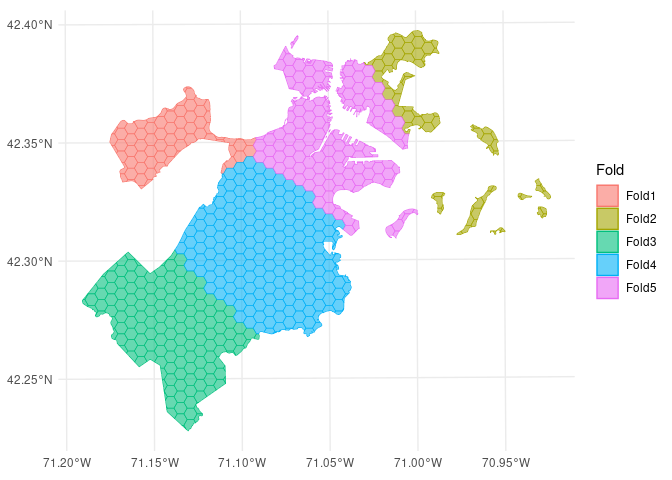
We can use the same function to visualize each fold separately:
library(purrr)
walk(folds$splits, function(x) print(autoplot(x)))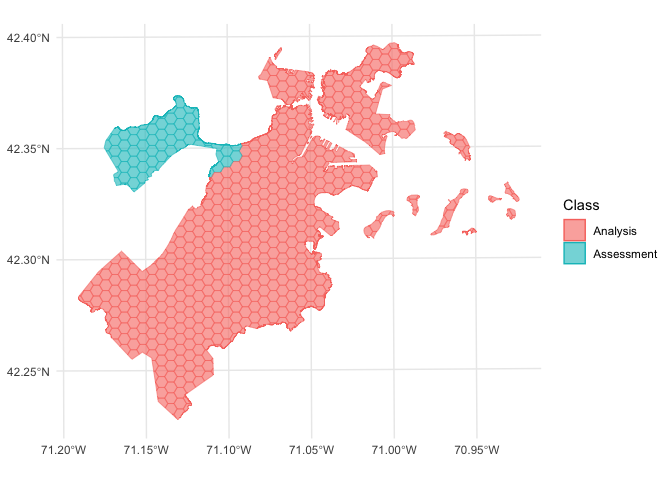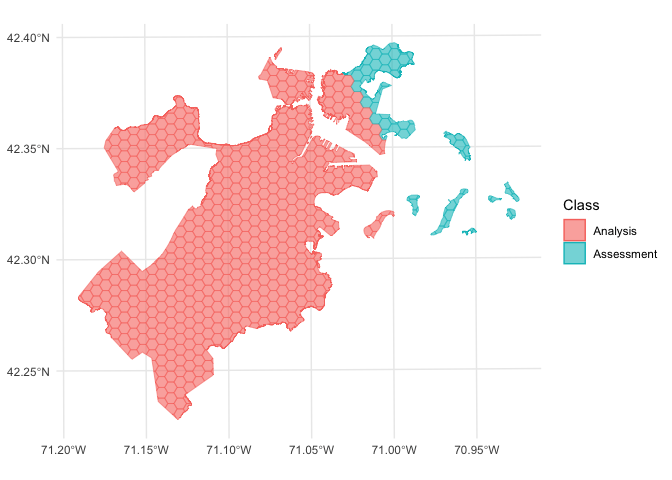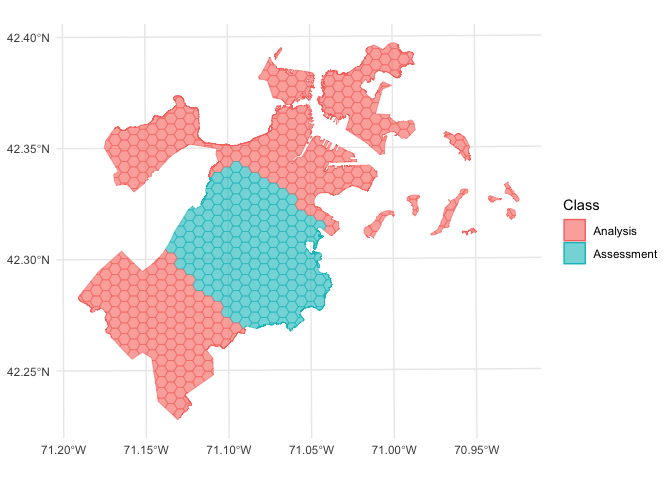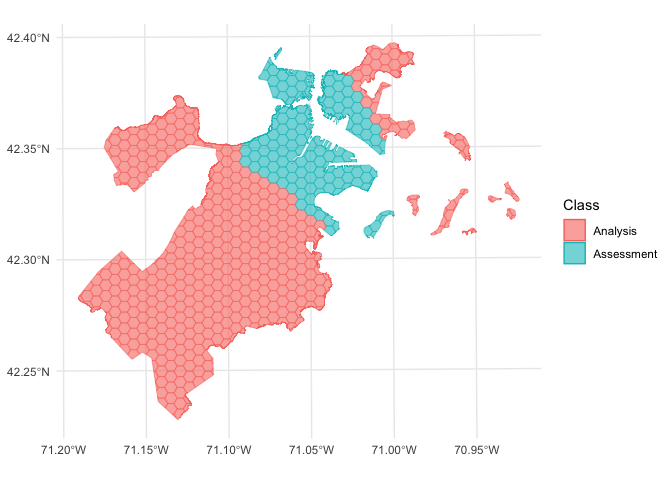
So far, we’ve only scratched the surface of the functionality spatialsample provides. For more information, check out the Getting Started documentation!
This project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
For questions and discussions about tidymodels packages, modeling, and machine learning, please post on RStudio Community.
If you think you have encountered a bug, please submit an issue.
Either way, learn how to create and share a reprex (a minimal, reproducible example), to clearly communicate about your code.
Check out further details on contributing guidelines for tidymodels packages and how to get help.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.