The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
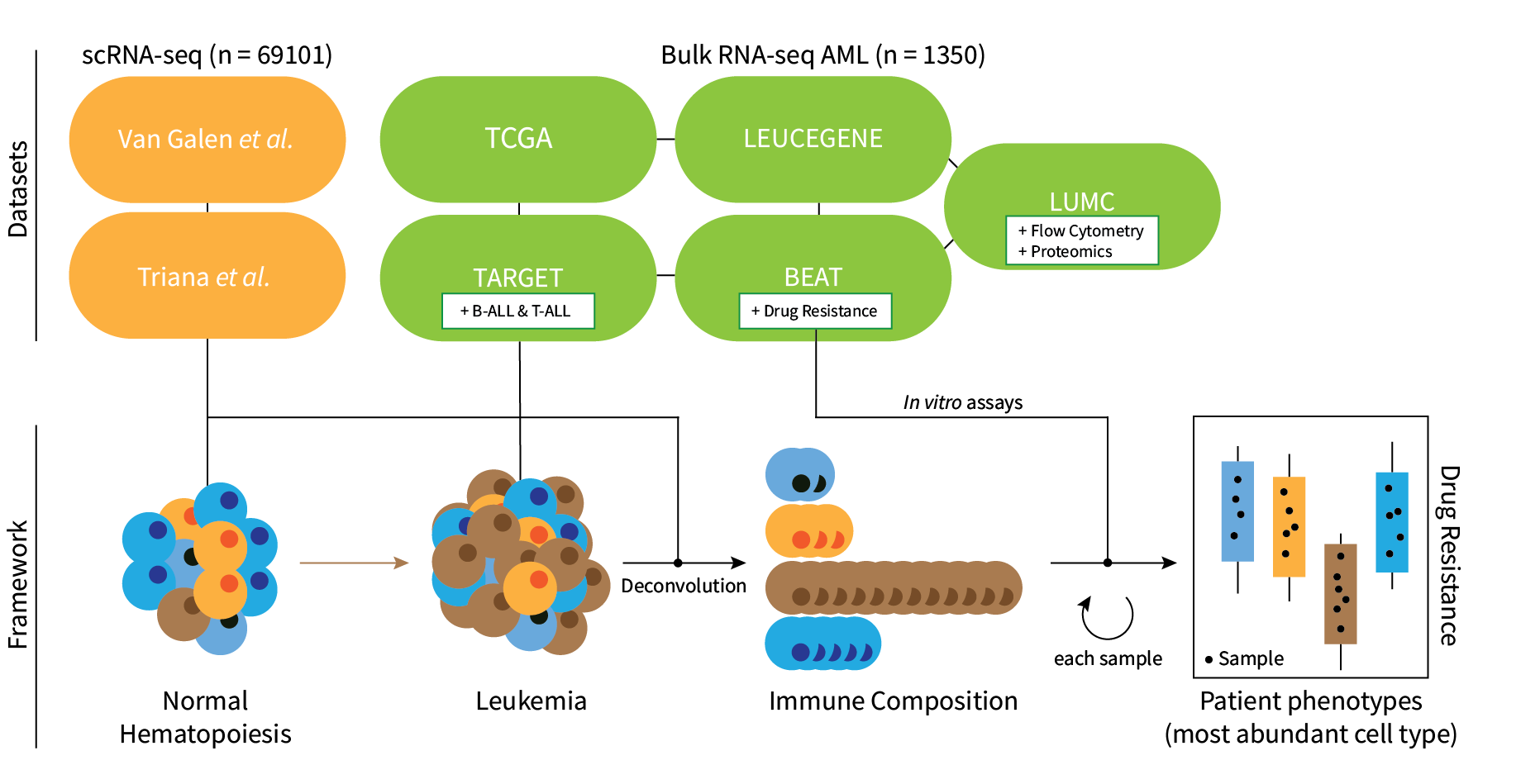
seAMLess is a wrapper function which deconvolutes bulk
Acute Myeloid Leukemia (AML) RNA-seq samples with a healthy single cell
reference atlas.

# CRAN mirror
install.packages("seAMLess")To get bug fix and use a feature from the development version:
# install.packages("devtools")
devtools::install_github("eonurk/seAMLess")
seAMLess is also available in Bioconda and can be
installed via:
conda install -c bioconda r-seamlessNext, install the seAMLessData package inside of R, this
will take a few minutes:
install.packages("seAMLessData", repos = "https://eonurk.github.io/drat/")library(seAMLess)
library(xbioc) # required
data(exampleTCGA)
head(exampleTCGA)[,1:4]## X TCGA.AB.2856.03A TCGA.AB.2849.03A TCGA.AB.2971.03A
## 1 ENSG00000000003.13 7 9 1
## 2 ENSG00000000005.5 0 1 0
## 3 ENSG00000000419.11 689 661 555
## 4 ENSG00000000457.12 633 1434 855
## 5 ENSG00000000460.15 372 1211 519
## 6 ENSG00000000938.11 14712 405 6076# Now run the function
res <- seAMLess(exampleTCGA)## >> Human ensembl ids are converted to symbols...
## >> Deconvoluting samples...
## >> Deconvolution completed...
## >> Predicting Venetoclax resistance...
## >> Done...# AML deconvolution
head(res$Deconvolution)[,1:4]## CD14 Mono GMP T Cells pre B
## TCGA.AB.2856.03A 0.1495886 0.7079107 0.022868623 0.00000000
## TCGA.AB.2849.03A 0.0000000 0.0000000 0.000000000 0.00000000
## TCGA.AB.2971.03A 0.5494418 0.4462562 0.002898678 0.00000000
## TCGA.AB.2930.03A 0.0000000 0.4698555 0.000000000 0.00000000
## TCGA.AB.2891.03A 0.0000000 0.6189622 0.018645484 0.01384012
## TCGA.AB.2872.03A 0.0000000 0.9950150 0.000000000 0.00000000# Create ternary plot
ternaryPlot(res)
# Venetoclax resistance
res$Venetoclax.resistance[1:4]## TCGA.AB.2856.03A TCGA.AB.2849.03A TCGA.AB.2971.03A TCGA.AB.2930.03A
## 0.5070113 0.3242576 0.6678995 0.3305996You can send pull requests to make your contributions.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.