The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Ridson Al Farizal P, Azka Ubaidillah
Ridson Al Farizal P ridsonap@bps.go.id
Implementation of small area estimation (Fay-Herriot model) with EBLUP (Empirical Best Linear Unbiased Prediction) Approach for non-sampled area estimation by adding cluster information and assuming that there are similarities among particular areas. See also Rao & Molina (2015, ISBN:978-1-118-73578-7) and Anisa et al. (2013) doi:10.9790/5728-10121519.
You can install the development version of saens from GitHub with:
# install.packages("devtools")
devtools::install_github("Alfrzlp/saens")or you can install cran version
install.packages("saens")This is a basic example which shows you how to solve a common problem:
library(saens)
library(sae)
library(dplyr)
library(tidyr)
library(ggplot2)
windowsFonts(
poppins = windowsFont('poppins')
)# Load data set from sae package
data(milk)
milk$var <- milk$SD^2
glimpse(mys)
#> Rows: 42
#> Columns: 8
#> $ area <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 1…
#> $ y <dbl> 8.359527, 7.599650, 5.514137, 3.869326, 6.305063, 3.926807, 5.68…
#> $ var <dbl> 0.6618838, 0.8374691, 0.8822257, 0.6581716, 1.2788021, 0.3878004…
#> $ rse <dbl> 9.732158, 12.041783, 17.033831, 20.966899, 17.935449, 15.858590,…
#> $ x1 <dbl> 124, 89, 57, 88, 141, 96, 146, 110, 58, 63, 35, 56, 84, 240, 123…
#> $ x2 <dbl> 24, 18, 19, 35, 46, 29, 57, 41, 18, 13, 12, 14, 38, 71, 33, 53, …
#> $ x3 <dbl> 14, 9, 5, 19, 29, 10, 34, 23, 11, 5, 9, 11, 35, 48, 29, 44, 37, …
#> $ clust <dbl> 3, 3, 4, 4, 3, 4, 3, 3, 3, 3, 3, 3, 3, 4, 3, 3, 3, 3, 4, 3, 3, 3…
glimpse(milk)
#> Rows: 43
#> Columns: 7
#> $ SmallArea <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 1…
#> $ ni <int> 191, 633, 597, 221, 195, 191, 183, 188, 204, 188, 149, 290, …
#> $ yi <dbl> 1.099, 1.075, 1.105, 0.628, 0.753, 0.981, 1.257, 1.095, 1.40…
#> $ SD <dbl> 0.163, 0.080, 0.083, 0.109, 0.119, 0.141, 0.202, 0.127, 0.16…
#> $ CV <dbl> 0.148, 0.074, 0.075, 0.174, 0.158, 0.144, 0.161, 0.116, 0.12…
#> $ MajorArea <int> 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, …
#> $ var <dbl> 0.026569, 0.006400, 0.006889, 0.011881, 0.014161, 0.019881, …model1 <- eblupfh(yi ~ as.factor(MajorArea), data = milk, vardir = "var")
#> ✔ Convergence after 4 iterations
#> → Method : REML
#>
#> ── Coefficient ─────────────────────────────────────────────────────────────────
#> beta Std.Error z-value p-value
#> (Intercept) 0.968189 0.069362 13.9585 < 2.2e-16 ***
#> as.factor(MajorArea)2 0.132780 0.103001 1.2891 0.197357
#> as.factor(MajorArea)3 0.226946 0.092330 2.4580 0.013972 *
#> as.factor(MajorArea)4 -0.241301 0.081617 -2.9565 0.003111 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1AIC(model1)
#> AIC
#> -15.35496
BIC(model1)
#> BIC
#> -6.548956
logLik(model1)
#> loglike
#> 12.67748coef(model1)
#> (Intercept) as.factor(MajorArea)2 as.factor(MajorArea)3
#> 0.9681890 0.1327801 0.2269462
#> as.factor(MajorArea)4
#> -0.2413011summary(model1)
#> beta Std.Error z-value p-value
#> (Intercept) 0.968189 0.069362 13.9585 < 2.2e-16 ***
#> as.factor(MajorArea)2 0.132780 0.103001 1.2891 0.197357
#> as.factor(MajorArea)3 0.226946 0.092330 2.4580 0.013972 *
#> as.factor(MajorArea)4 -0.241301 0.081617 -2.9565 0.003111 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1saens::autoplot(model1, variable = 'MSE')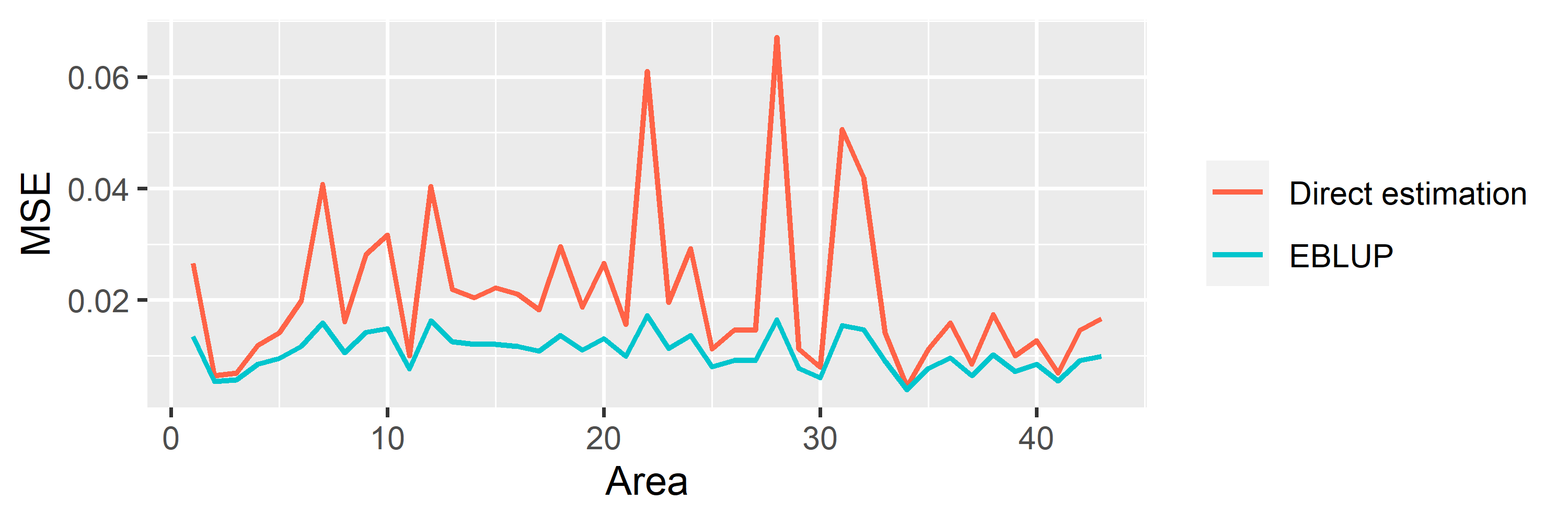
saens::autoplot(model1, variable = 'RRMSE')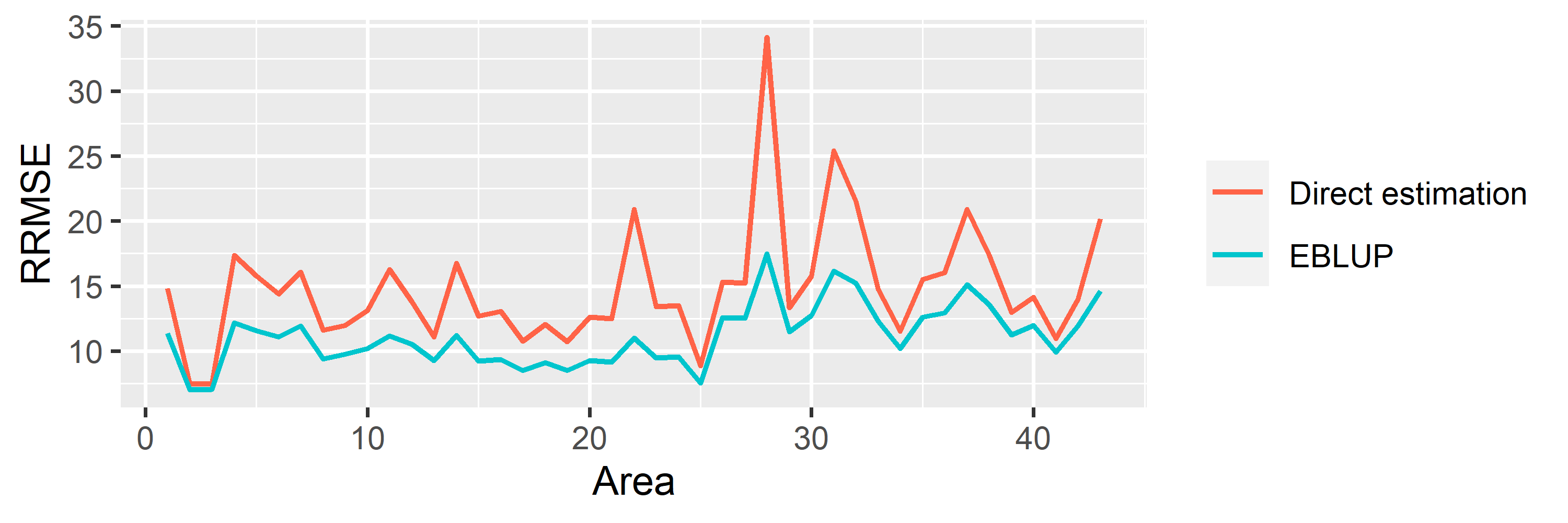
saens::autoplot(model1, variable = 'estimation')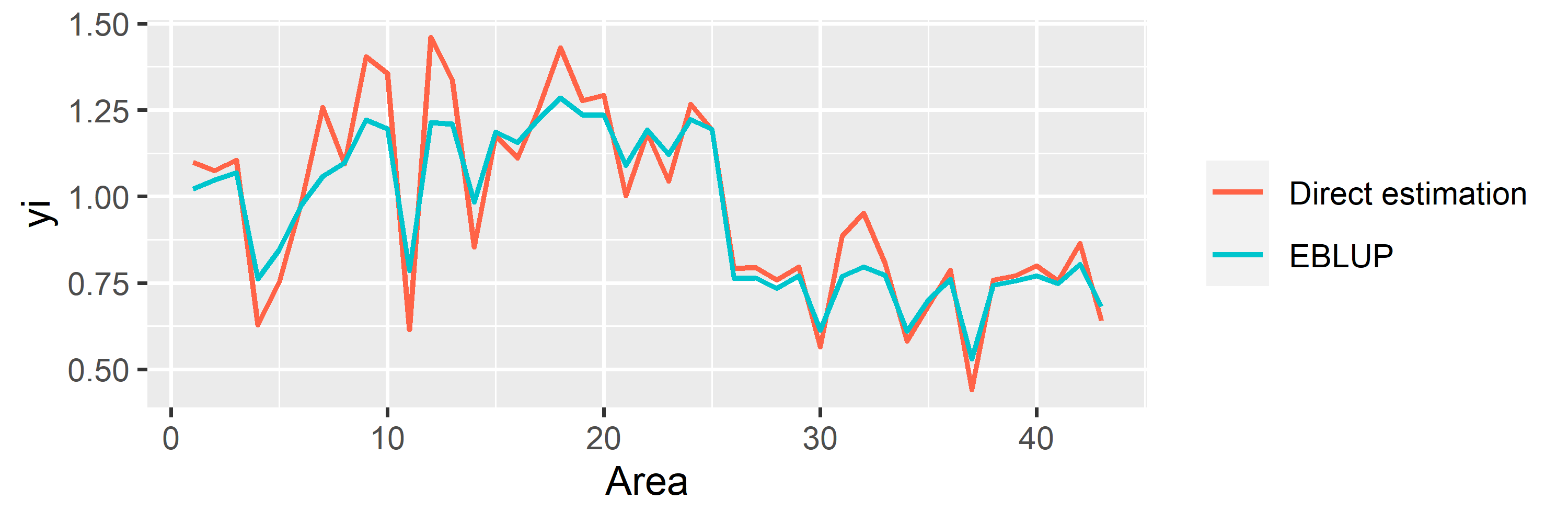
model_ns <- eblupfh_cluster(y ~ x1 + x2 + x3, data = mys, vardir = "var", cluster = "clust")
#> ✔ Convergence after 6 iterations
#> → Method : REML
#>
#> ── Coefficient ─────────────────────────────────────────────────────────────────
#> beta Std.Error z-value p-value
#> (Intercept) 3.1077510 0.7697687 4.0373 5.408e-05 ***
#> x1 -0.0019323 0.0098886 -0.1954 0.8451
#> x2 0.0555184 0.0614129 0.9040 0.3660
#> x3 0.0335344 0.0580013 0.5782 0.5632
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1mys$eblup_est <- model_ns$df_res$eblup
mys$eblup_rse <- model_ns$df_res$rse
glimpse(mys)
#> Rows: 42
#> Columns: 10
#> $ area <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 1…
#> $ y <dbl> 8.359527, 7.599650, 5.514137, 3.869326, 6.305063, 3.926807, …
#> $ var <dbl> 0.6618838, 0.8374691, 0.8822257, 0.6581716, 1.2788021, 0.387…
#> $ rse <dbl> 9.732158, 12.041783, 17.033831, 20.966899, 17.935449, 15.858…
#> $ x1 <dbl> 124, 89, 57, 88, 141, 96, 146, 110, 58, 63, 35, 56, 84, 240,…
#> $ x2 <dbl> 24, 18, 19, 35, 46, 29, 57, 41, 18, 13, 12, 14, 38, 71, 33, …
#> $ x3 <dbl> 14, 9, 5, 19, 29, 10, 34, 23, 11, 5, 9, 11, 35, 48, 29, 44, …
#> $ clust <dbl> 3, 3, 4, 4, 3, 4, 3, 3, 3, 3, 3, 3, 3, 4, 3, 3, 3, 3, 4, 3, …
#> $ eblup_est <dbl> 7.612738, 6.782316, 5.187060, 4.201545, 6.323679, 4.048590, …
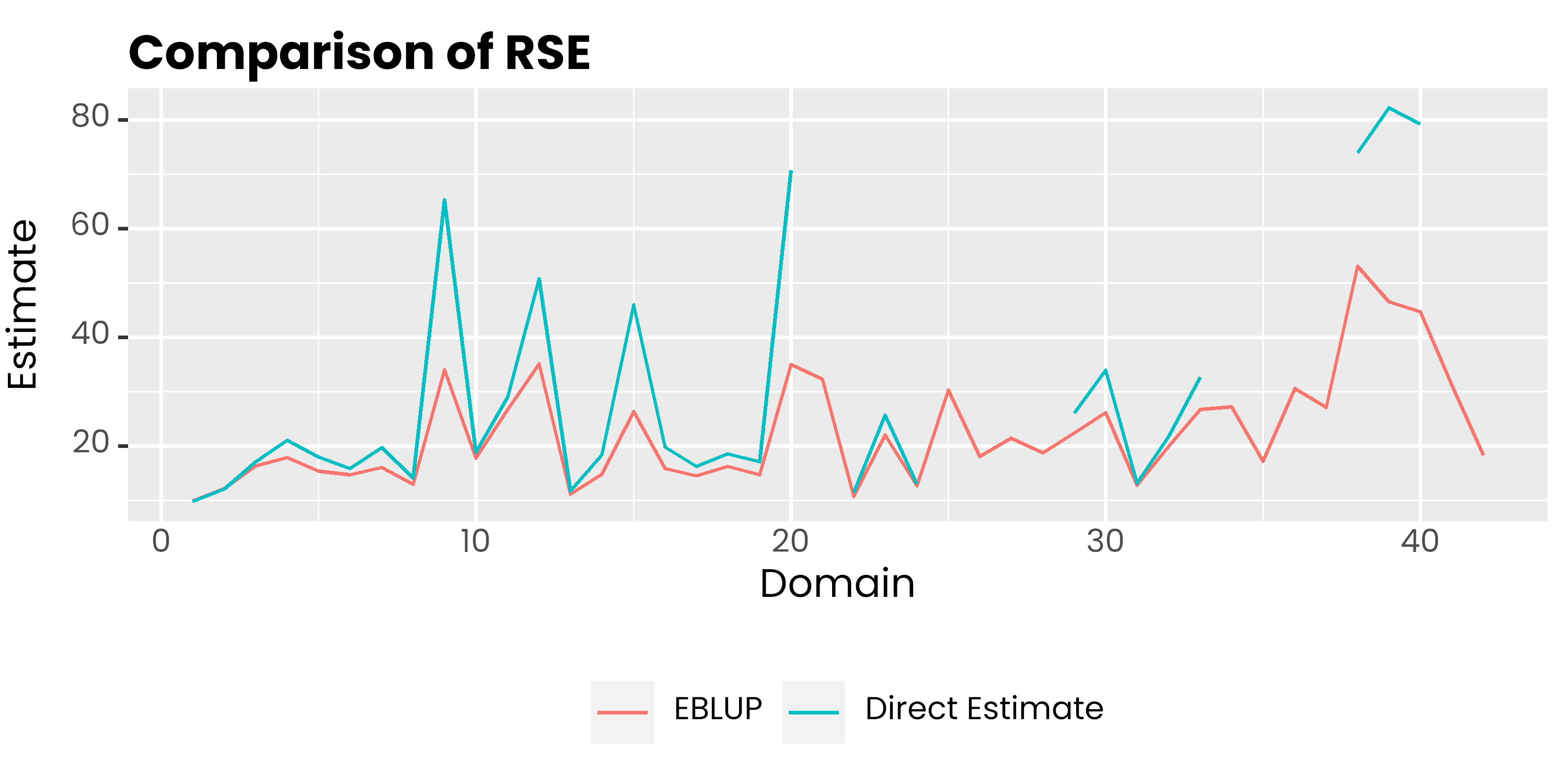
#> $ eblup_rse <dbl> 9.844182, 12.144869, 16.347620, 17.841554, 15.307573, 14.713…mys %>%
select(area, rse, eblup_rse) %>%
pivot_longer(-1, names_to = "metode", values_to = "rse") %>%
ggplot(aes(x = area, y = rse, col = metode)) +
geom_line() +
scale_color_discrete(
labels = c('EBLUP', 'Direct Estimate')
) +
labs(col = NULL, y = 'Estimate', x = 'Domain', title = 'Comparison of RSE') +
theme(
legend.position = 'bottom',
text = element_text(family = 'poppins'),
axis.ticks.x = element_blank(),
plot.title = element_text(face = 2, vjust = 0),
plot.subtitle = element_text(colour = 'gray30', vjust = 0)
)
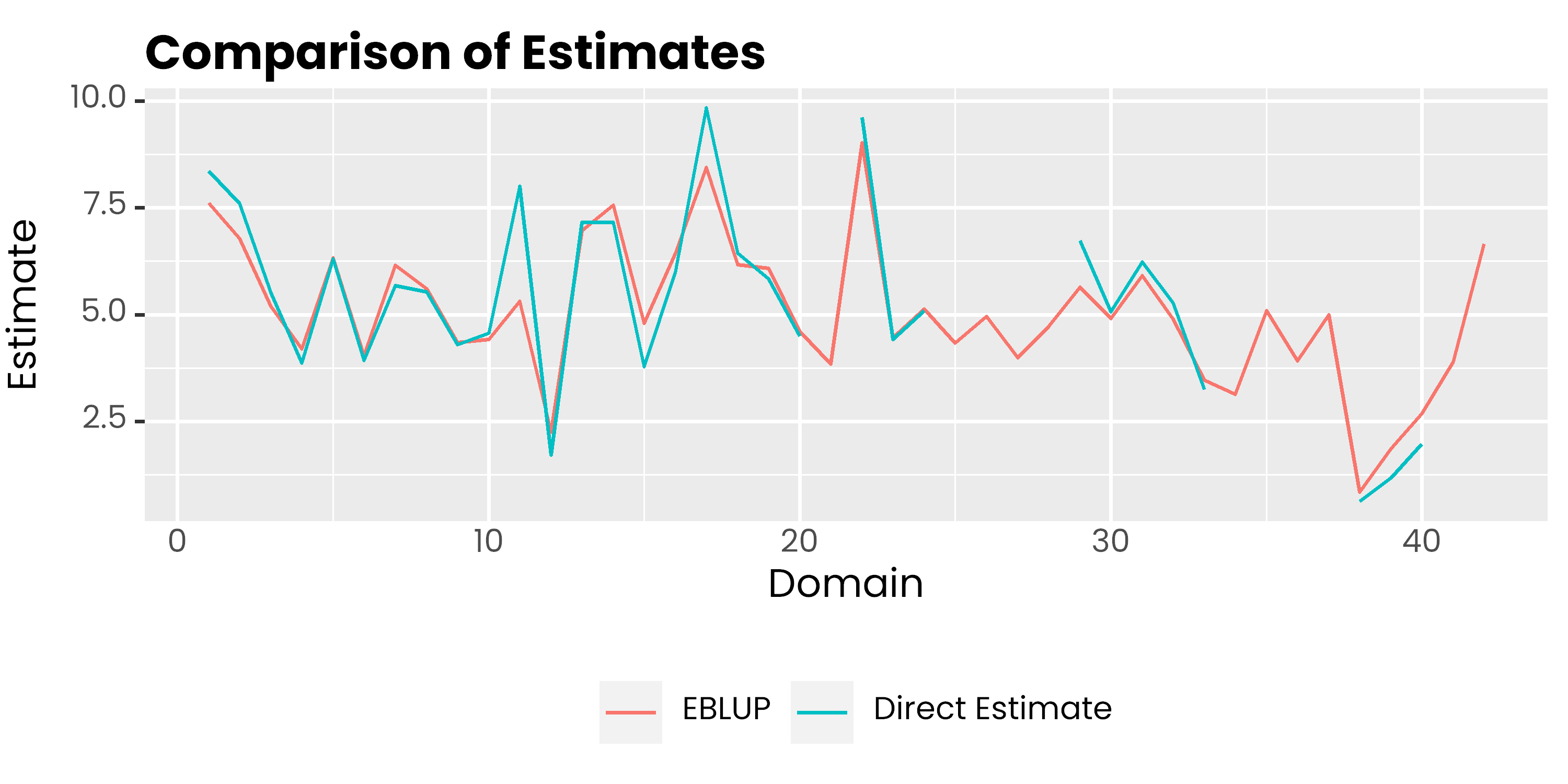
mys %>%
select(area, y, eblup_est) %>%
pivot_longer(-1, names_to = "metode", values_to = "rse") %>%
ggplot(aes(x = area, y = rse, col = metode)) +
geom_line() +
scale_color_discrete(
labels = c('EBLUP', 'Direct Estimate')
) +
labs(col = NULL, y = 'Estimate', x = 'Domain', title = 'Comparison of Estimates') +
theme(
legend.position = 'bottom',
text = element_text(family = 'poppins'),
axis.ticks.x = element_blank(),
plot.title = element_text(face = 2, vjust = 0),
plot.subtitle = element_text(colour = 'gray30', vjust = 0)
)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.