The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

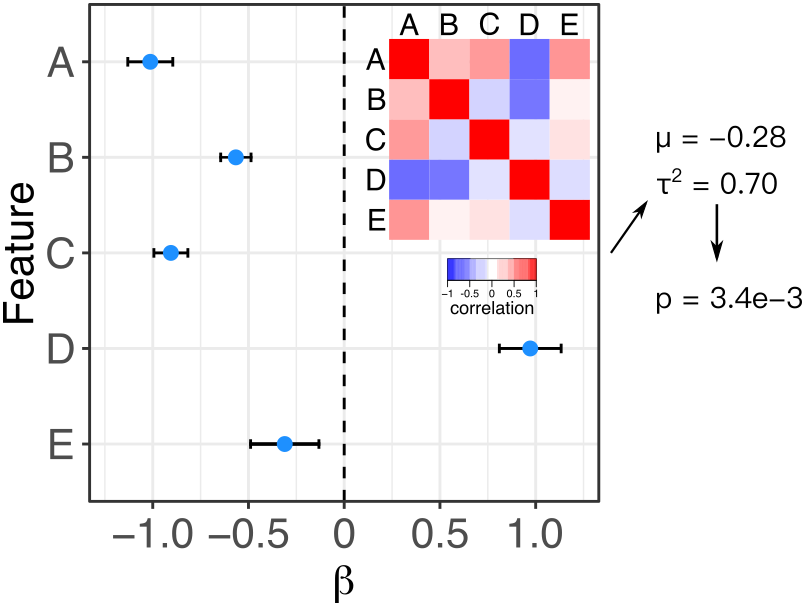
Meta-analysis is widely used to summarize estimated effects sizes across multiple statistical tests. Standard fixed and random effect meta-analysis methods assume that the estimated of the effect sizes are statistically independent. Here we relax this assumption and enable meta-analysis when the correlation matrix between effect size estimates is known. Fixed effect meta-analysis uses the method of Lin and Sullivan (2009), and random effects meta-analysis uses the method of Han, et al. 2016. An exentsion of the Lin-Sullivan method for finite sample size is described in Hoffman and Roussos (2025).
# Run fixed effects meta-analysis,
# accounting for correlation
LS( beta, stders, Sigma)
# Run fixed effects meta-analysis,
# accounting for correlation,
# and finite sample size using residual degrees of freedom
LS.empirical( beta, stders, Sigma, nu=rdf)
# Run random effects meta-analysis,
# accounting for correlation
RE2C( beta, stders, Sigma)devtools::install_github("DiseaseNeurogenomics/remaCor")These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.