The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Comparison of color maps produced using k-means clustering (center) vs.
recolorize(right). Original image credits: John E. Randall, Nathan P. Lord, and Alison Davis-Rabosky.
This is a package for making color maps, which are needed (or at least useful) for a wide range of color analysis techniques. It was born out of conversations with many biologists who found, to their surprise and mine, that generating color maps were the bottleneck step in their analyses. Fully automated methods rarely work all of the time, and are difficult to modify, while fully manual methods are subjective and time-consuming. This package tries to split the difference by giving you a mix of tools that will do a pretty good job with no user input, and then allow minor manual changes like merging and filtering layers or splitting components, before exporting them to the next step of your analysis (e.g. pavo, patternize). It’s also, for the most part, totally deterministic – no arbitrary seed-setting for repeatability.
Install the package:
# development version:
install.packages("devtools")
devtools::install_github("hiweller/recolorize")
# OR
install.packages("recolorize") # CRAN releaseTo run recolorize on a single image:
library(recolorize)
# load an image that comes with the package:
img <- system.file("extdata/corbetti.png", package = "recolorize")
rc <- recolorize2(img, cutoff = 45)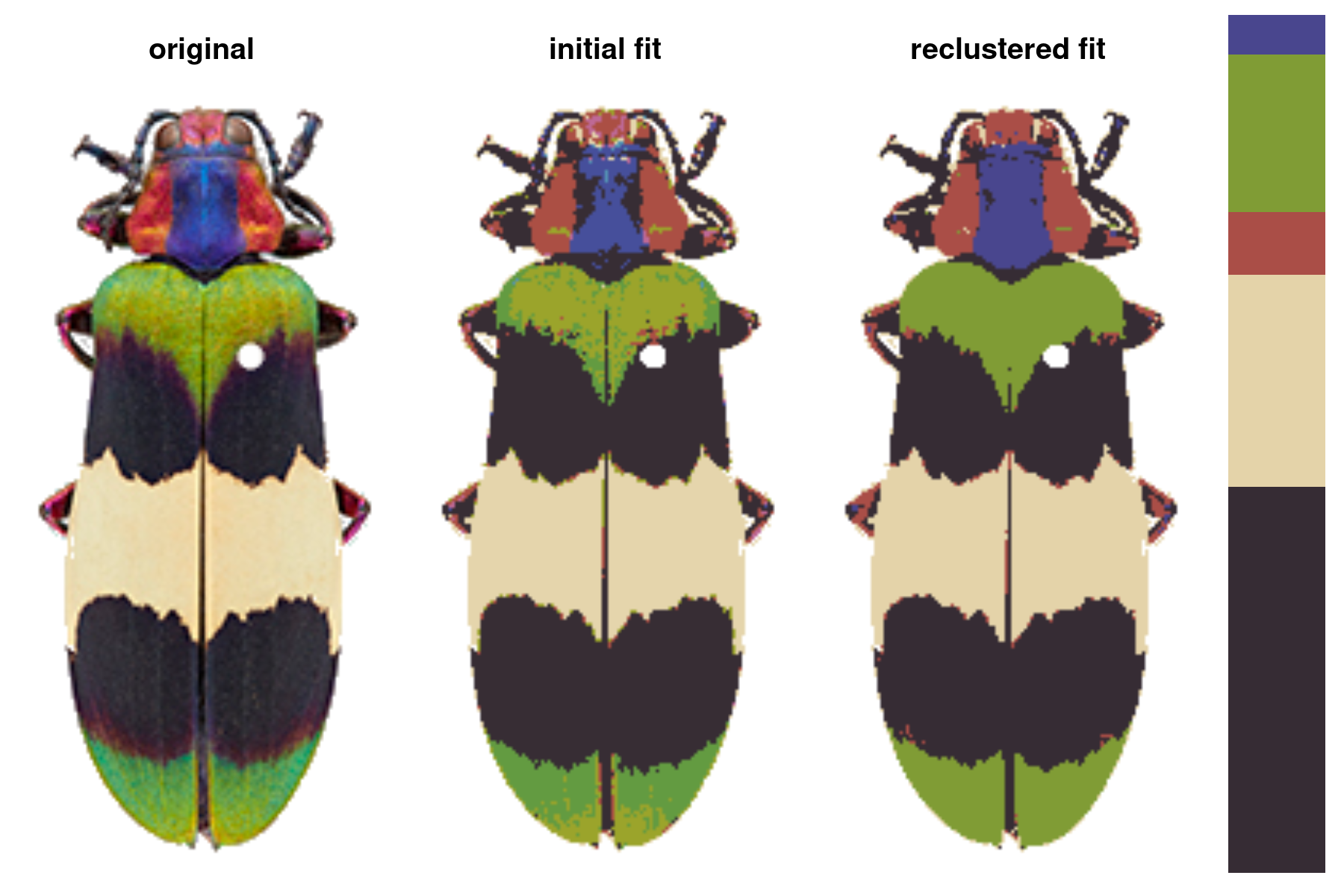
A batch processing example:
# get list of all PNGs that come with the package:
images <- dir(system.file("extdata", package = "recolorize"),
pattern = ".png", full.names = TRUE)
# for every image...
for (i in 1:length(images)) {
# get an initial fit with generic clustering
init_fit <- recolorize2(images[i], method = "hist", bins = 2, cutoff = 25, plotting = FALSE)
# drop small patches
refined_fit <- thresholdRecolor(init_fit, pct = 0.01, plotting = FALSE)
# store in an output variable
if (i == 1) {
colormap_list <- list(refined_fit)
} else {
colormap_list[[i]] <- refined_fit
}
}
# compare original to recolored images:
layout(matrix(1:10, nrow = 2, byrow = TRUE))
par(mar = rep(0, 4))
o <- lapply(colormap_list, function(i) plot(i$original_img))
r <- lapply(colormap_list, function(i) plotImageArray(recoloredImage(i)))
See package vignettes for detailed documentation.
Recolorize is a toolbox of automatic, semi-automatic, and manual methods that aims to give you reproducible results that work pretty well out-of-box, and which are easy to tailor to your needs. The general workflow is an initial clustering step -> an automatic or semi-automatic reclustering step -> optional manual refinements -> export to another format, like an image, a set of binary masks, or another R package.
Please do email me if this is something that may be relevant for you or if you have questions. Most of the features in the package are the result of feedback, and no method should be developed in a vacuum. If you include example images in your email, my likelihood of responding goes through the roof.
Email: hannahiweller@gmail.com
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.