The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Simulates the dynamics of exploited fish populations using the Jones modification of the Beverton-Holt equilibrium yield equation to compute yield-per-recruit and dynamic pool models (Ricker 1975) https://publications.gc.ca/site/eng/480738/publication.html. Allows users to evaluate minimum, slot, and inverted length limits on exploited fisheries using specified life history parameters. Users can simulate population under a variety of conditional fishing mortality and conditional natural mortality. Calculated quantities include number of fish harvested and dying naturally, mean weight and length of fish harvested, number of fish that reach specified lengths of interest, total number of fish and biomass in the population, and stock density indices.
This project is supported in part by the American Fisheries Society Data and Technology Section (https://units.fisheries.org/fits/)
The package currently replicates all yield per recruit and dynamic pool modeling in FAMS with the exception of spawning potential ratio. Spawning potential ratio calculations will be added at a later date. Life history parameters can be estimated using FSA.
The most recent stable version from CRAN may be installed with
install.packages("rFAMS")The development version may be installed from GitHub with
# install.packages("devtools")
devtools::install_github("fishR-Core-Team/rFAMS")You may need R Tools installed on your system to install the development version from GitHub. See the instructions for (R Tools for Windows or R Tools for Mac OS X).
Report questions, comments, or bug reports on the issues page.
Please adhere to the Code of Conduct.
This is a basic example which shows you how simulate yield based on a single minimum length limit with variable cf and variable cm:
#Load other required packages for organizing output and plotting
library(rFAMS)
#> ## rFAMS v0.0.2. See citation('rFAMS') if used in publication.
#> ## Visit https://github.com/fishR-Core-Team/rFAMS/issues to report any bugs.
library(tidyr) ## for pivot_longer
library(dplyr) ## for filter
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(ggplot2) ## for ggplot et al.
# Life history parameters to be used below
LH <- makeLH(N0=100,tmax=15,Linf=592,K=0.20,t0=-0.3,LWalpha=-5.528,LWbeta=3.273)
# Estimate yield for one minLL and multiple cf and cm values.
# This is a minimal example, lengthinc, cfinc, cminc would likely be smaller
# to produce finer-scaled results
Res_1 <- yprBH_minLL_fixed(minLL=200,
cfmin=0.1,cfmax=0.9,cfinc=0.1,
cmmin=0.1,cmmax=0.9,cminc=0.1,
loi=c(200,250,300,350),lhparms=LH)
# Custom theme for plots (to make look nice)
theme_FAMS <- function(...) {
theme_bw() +
theme(
panel.grid.major=element_blank(),panel.grid.minor=element_blank(),
axis.text=element_text(size=14,color="black"),
axis.title=element_text(size=16,color="black"),
axis.title.y=element_text(angle=90),
axis.line=element_line(color="black"),
panel.border=element_blank()
)
}
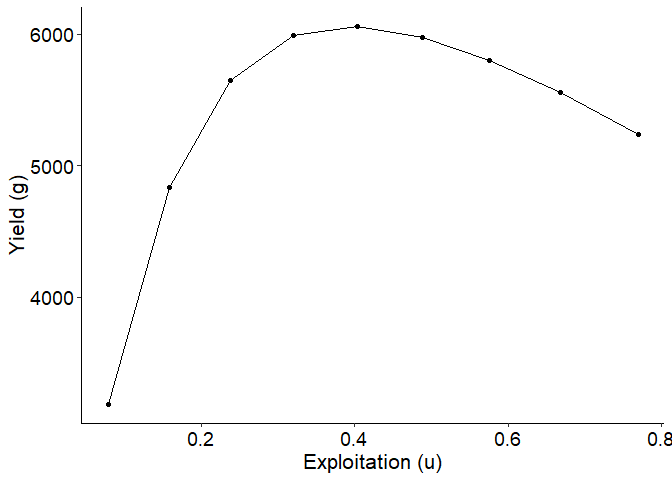
# Yield curve (yield vs exploitation)
# Extract results for cm=0.40
plot_dat <- Res_1 |> dplyr::filter(cm==0.40)
ggplot(data=plot_dat,mapping=aes(x=u,y=yield)) +
geom_point() +
geom_line() +
labs(y="Yield (g)",x="Exploitation (u)") +
theme_FAMS()
#Plot number of fish reaching 300 mm as a function of exploitation with cm = 0.40
ggplot(data=plot_dat,mapping=aes(x=u,y=`N at 300 mm`)) +
geom_point() +
geom_line() +
labs(y="Number of fish at 300 mm",x="Exploitation (u)") +
theme_FAMS()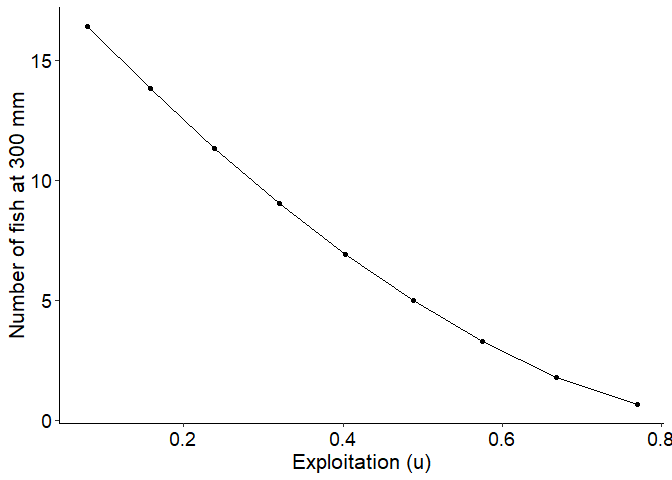
# Plot number of fish reaching 300 mm as a function of exploitation with cm = 0.40
# Select columns for plotting and convert to long
plot_data_long <- plot_dat %>%
select(u,`N at 200 mm`, `N at 250 mm`, `N at 300 mm`, `N at 350 mm`) %>%
pivot_longer(!u, names_to="loi",values_to="number")
# Generate plot
ggplot(data=plot_data_long,mapping=aes(x=u,y=number,group=loi,color=loi)) +
geom_point() +
scale_color_discrete(name="Yield",labels=c("N at 200 mm", "N at 250 mm", "N at 300 mm", "N at 350 mm"))+
geom_line() +
labs(y="Number of fish",x="Exploitation (u)") +
theme_FAMS() +
theme(legend.position = "top")+
guides(color=guide_legend(title="Length of interest"))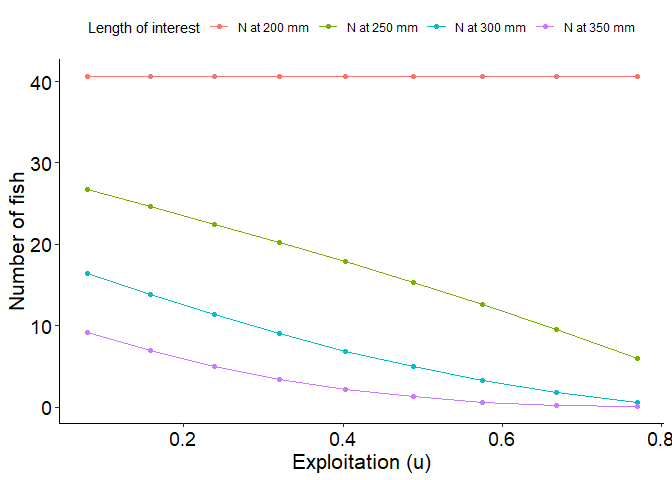
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.