
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

# Install in R
install.packages("quickcode")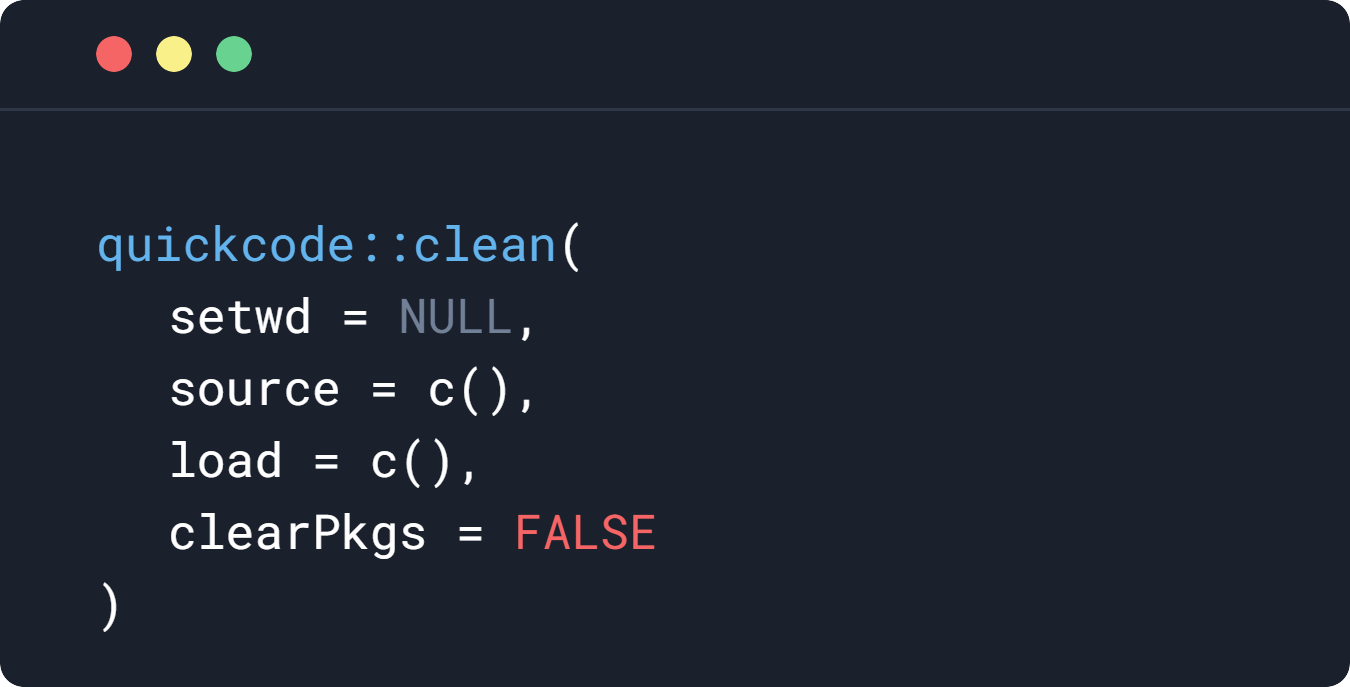


# Use the nullish coalescing operator using "or()" or "%or%"
ex.V1 <- 5
ex.V2 <- NA
ex.V3 <- NULL
ex.V4 <- ""
alternative <- 500
# Give an alternative result if the test is NULL NA or empty
or(ex.V1,alternative) # result will give 5 because ex.V1 is not NULL NA or empty
ex.V1 %or% alternative # result will give 5 because ex.V1 is not NULL NA or empty
ex.V2 %or% alternative # result will give 500 because ex.V2 is NA
ex.V3 %or% alternative # result will give 500 because ex.V3 is NULL
ex.V4 %or% alternative # result will give 500 because ex.V4 is empty
# Further chaining
ex.V2 %or% ex.V1 %or% alternative # result will be 5 because ex.V2 is NA but ex.V1 is 5
ex.V2 %or% ex.V3 %or% alternative # result will be 500 because ex.V2 is NA and ex.V3 is NULL
#load libraries and print names along with versions
quickcode::libraryAll(
dplyr,
r2resize,
ggplot2
)
#simple conversion between boolean types
#input type is "vector"
baba <- c(TRUE,"y","n","YES","yes",FALSE,"f","F","T","t")
as.boolean(baba,1) # return vector as Yes/No
as.boolean(baba,2) # return vector as TRUE/FALSE
as.boolean(baba,3) # return vector as 1/0
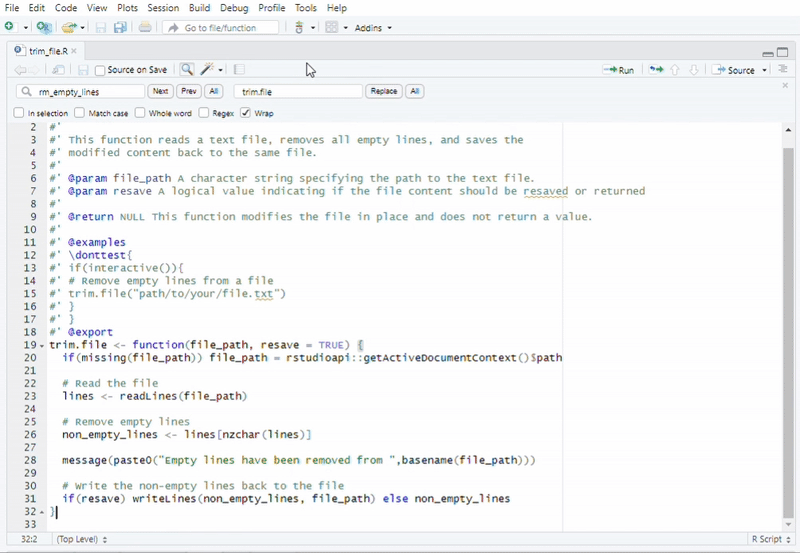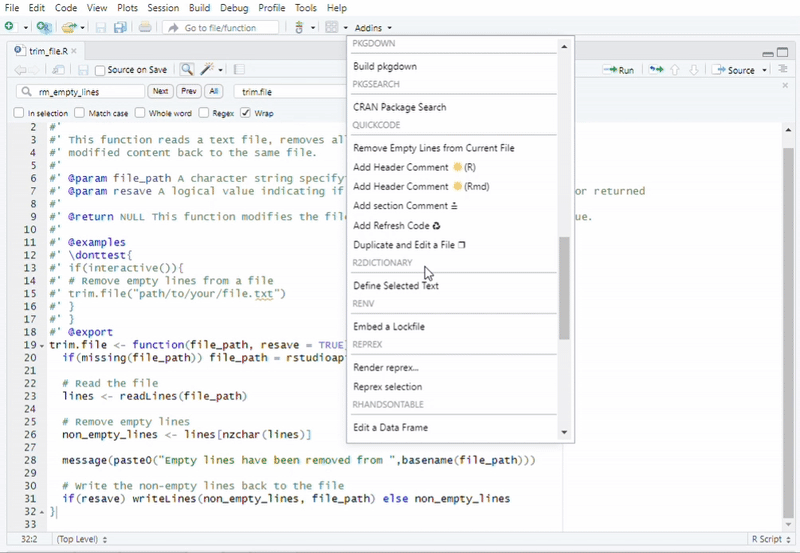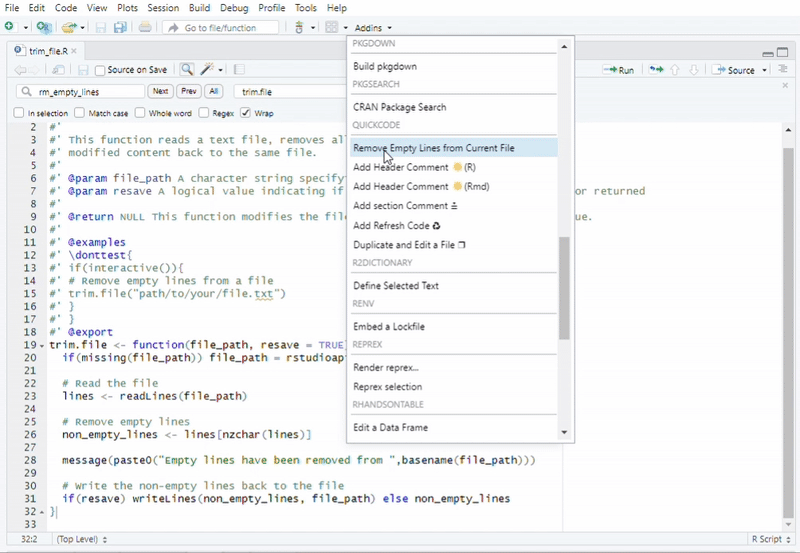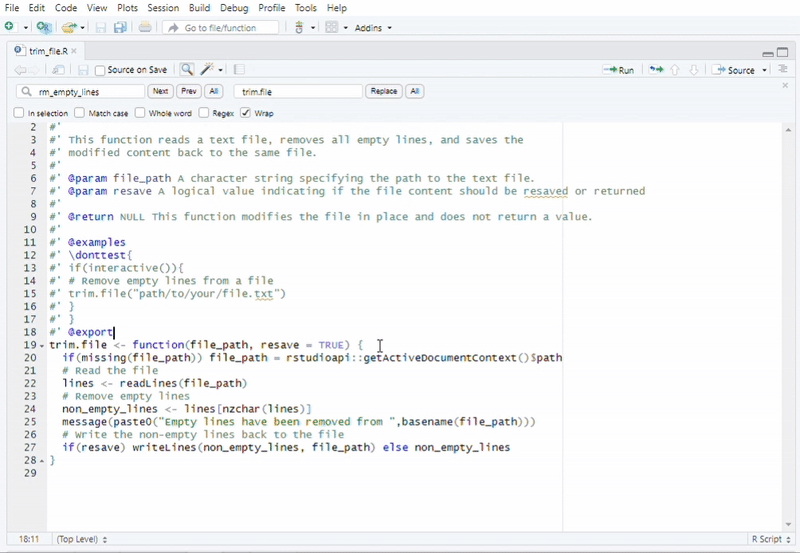
#Recommended to use "Addins" in RStudio
#Function call: Remove Empty Lines from a File
trim.file("path/to/your/file.txt")
#Detect outliers in data by IQR and Zscore methods
x <- c(1, 2, 3, 4, 100)
detect_outlier(x, summary = TRUE)
detect_outlier(z, method = "iqr", multiplier = 3)
detect_outlier(y, method = "zscore", z_threshold = 2.5)
#apply the yesNoBool to convert between boolean
#input type is "data.frame"
usedata <- data.frame(ID = number(32))
usedata #view the dataset
usedata$yess = rep(c("yes","n","no","YES","No","NO","yES","Y"),4) #create a new column
usedata #view the modified dataset
#set all yess field as standardize boolean
yesNoBool(usedata,yess, type="bin") #set all as binary 1/0
yesNoBool(usedata,yess, type="log") #set all as logical TRUE/FALSE
#initialize one or more variables
print(g) # Error: object 'g' not found
init(g,h,i,o)
print(g) # g = NULL
print(h) # h = NULL
init(r,y,u,b,value = 5)
print(r) # r = 5
print(b) # b = 5
print(z) # Error: object 'z' not found
#add keys to a vector content for use in downstream processes
ver1 <- c("Test 1","Test 2","Test 3")
add_key(ver1)
for(i in ver1){
message(sprintf("%s is the key for this %s", i$key, i$value))
}
# Introducing the super variable
# store dataset that should not be altered
newSuperVar(mtdf, value = austres) # create a super variable
head(mtdf) # view it
mtdf.class # view the store class of the variable, it cannot be changed
# it means that when the super variable is edited, the new value MUST have the same class
# create and lock super variable by default
# extra security to prevent changing
newSuperVar(mtdf3, value = beaver1, lock = TRUE)
head(mtdf3) # view
mtdf3.round(1) # round to 1 decimal places
head(mtdf3) # view
mtdf3.signif(2) # round to 2 significant digits
head(mtdf3) # view
# Task: create a new super variable to store numbers
# edit the numbers from various scopes
newSuperVar(edtvec, value = number(5))
edtvec # view content of the vector
# edtvec.set(letters) #ERROR: Cannot set to value with different class than initial value
edtvec.set(number(20)) # set to new numbers
edtvec # view output
for (pu in 1:8) {
print(edtvec) # view output within loop
edtvec.set(number(pu)) # set to new numbers within for loop
}
lc <- lapply(1:8, function(pu) {
print(edtvec) # view output within loop
edtvec.set(number(pu)) # set to new numbers within lapply loop
})
# see that the above changed the super variable easily.
# local variable will not be altered by the loop
# example
bim <- 198
lc <- lapply(1:8, function(j) {
print(bim)
bim <- j # will not alter the value of bim in next round
})
#check if the entry is not integer
not.integer(45) #returns TRUE
not.integer(45.) #returns TRUE
not.integer(45L) #returns FALSE
not.null(45L) #returns TRUE
not.null(h<-NULL) #returns FALSE
#clear R environment, set directory and load data
#note: the code below also automatically loads the quickcode library so that all other functions within package can be used easily
quickcode::refresh()
quickcode::clean()
#or combine with setwd and source and load
quickcode::clean(
setwd = "/wd/",
source = c(
"file.R",
"file2.R"
),
load = c(
"data.RData",
"data2.RData"
)
)
#shorthand for not in vector
p1 <- 4
p2 <- c(1:10)
p1 %nin% p2
#add to a vector in one code
p1 <- c(6,7,8)
p2 <- c(1,2,3)
vector_push(p1,p2)
print(p1)
#add to a data frame in one code
p1 <- data.frame(ID=1:10,ID2=1:10)
p2 <- data.frame(ID=11:20,ID2=21:30)
data_push(p1,p2,"rows")
print(p1)
#remove from a vector in one code
p1 <- c(6,7,8,1,2,3)
vector_pop(p1)
print(p1)
#remove from a data frame in one code
p1 <- data.frame(ID=1:10,ID2=1:10,CD=11:20,BD=21:30)
data_pop(p1) #remove last row
print(p1)
data_pop(p1,5) #remove last 5 rows
print(p1)
#remove columns from a data frame in one code
p1 <- data.frame(ID=1:10,ID2=1:10,ID4=1:10,CD=11:20,BD=21:30)
data_pop(p1,which = "cols") #remove last column
print(p1)
data_pop(p1,2,which = "cols") #remove last 2 columns
print(p1)
data_pop(p1,1,which = "cols") #remove last 1 column and vectorise
print(p1)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.