
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This package creates plots using quantile binning. Quantile binning is an exploratory data analysis tool that helps to see the distribution of the variables in a dataset as a function of the variable that is binned.
You can install the development version of qbinplots
from GitHub with:
remotes::install_github("edwindj/qbinplots")library(qbinplots)
## basic example codeA quantile binning boxplot
qbin_boxplot(iris, "Sepal.Length", n = 12)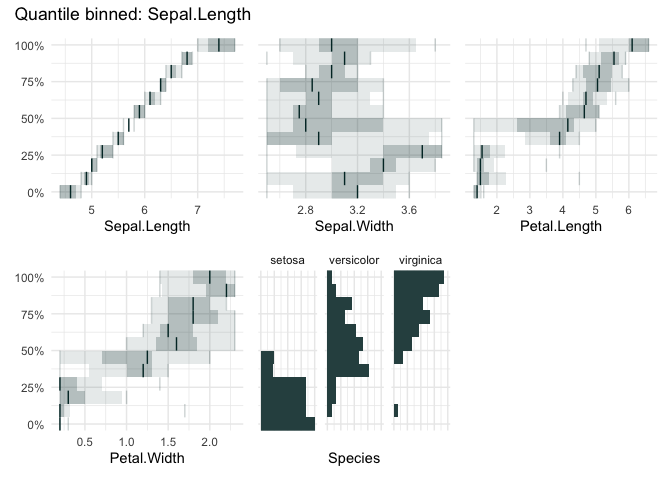
vs
A quantile binning barplot
qbin_barplot(iris, "Sepal.Length", n = 12)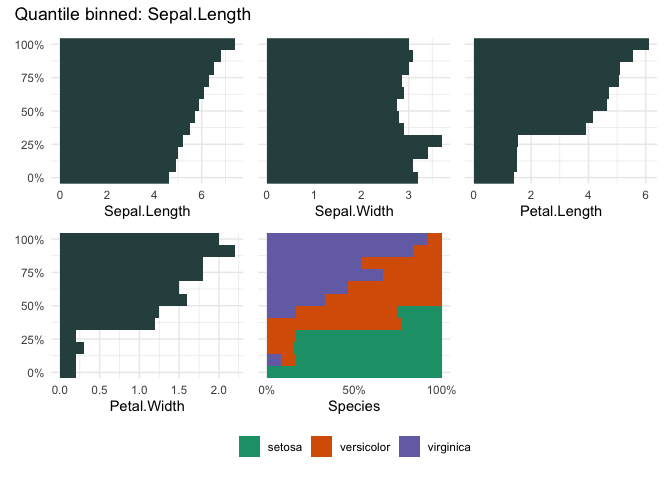
table_plot(iris, "Sepal.Length", n=12)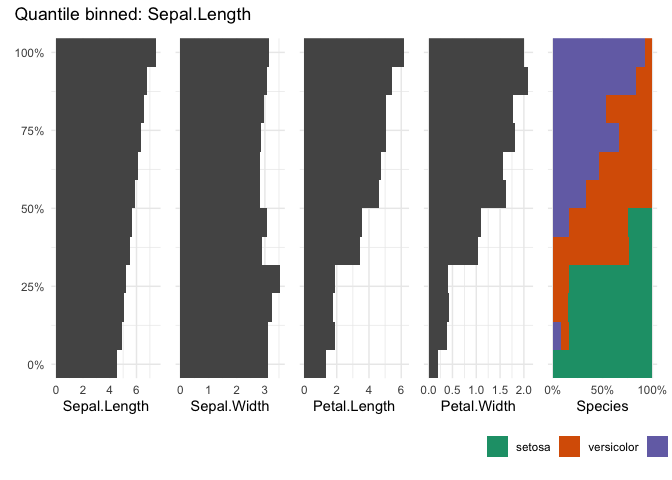
vs
A quantile binning heatmap
qbin_heatmap(iris, "Sepal.Length", n=12, auto_fill = TRUE)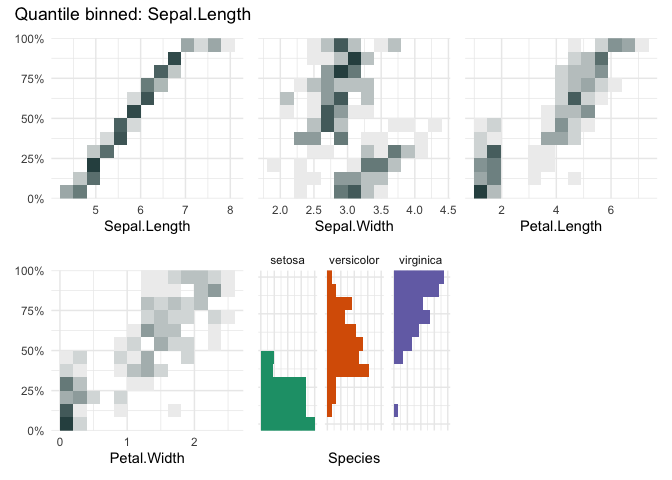
cond_boxplot(iris, "Sepal.Length", n=12, auto_fill = TRUE)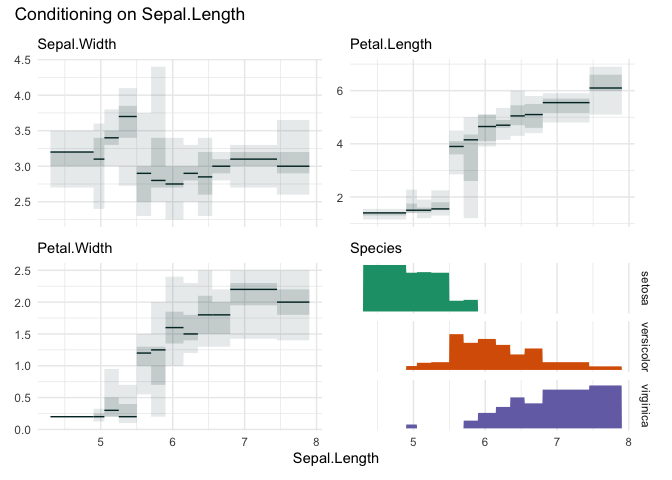
cond_barplot(iris, "Sepal.Length", n=12, auto_fill = TRUE)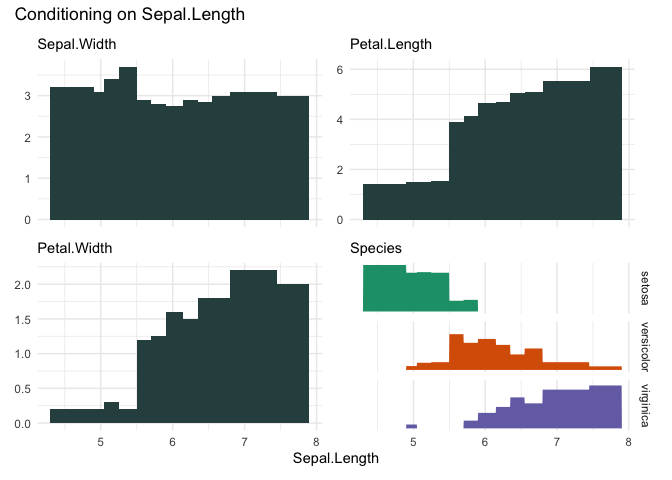
funq_plot(iris, "Sepal.Length", 12, auto_fill = TRUE)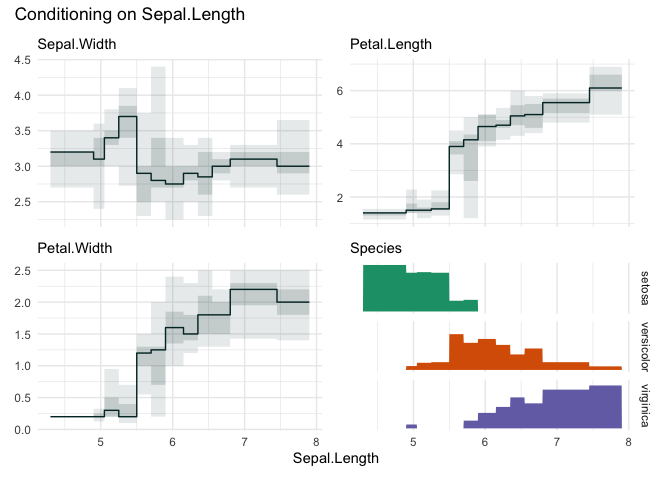
funq_plot(iris, "Sepal.Length", overlap = TRUE, min_bin_size = 20)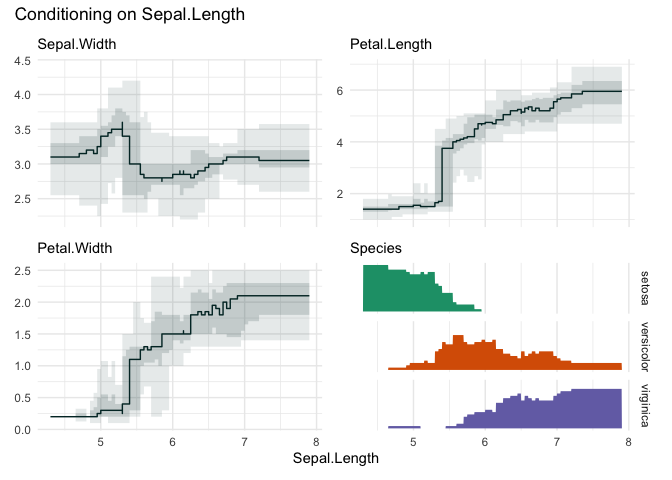
Choosing “Petal.Width”
funq_plot(iris, "Petal.Width", n=12)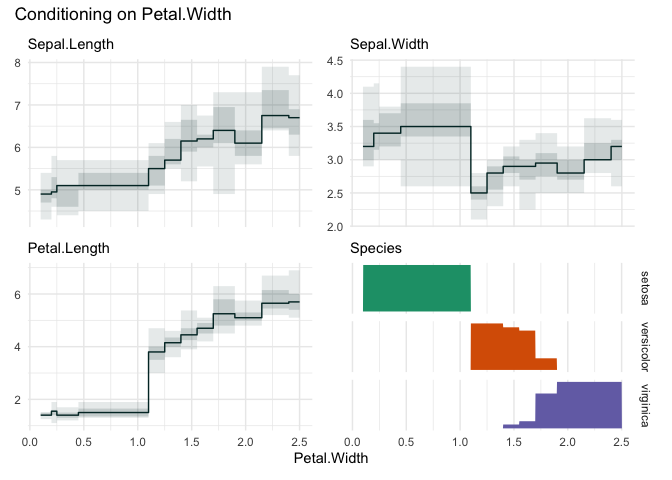
library(palmerpenguins)
qbin_lineplot(penguins[1:7], x="body_mass_g", n = 19, ncols = 4)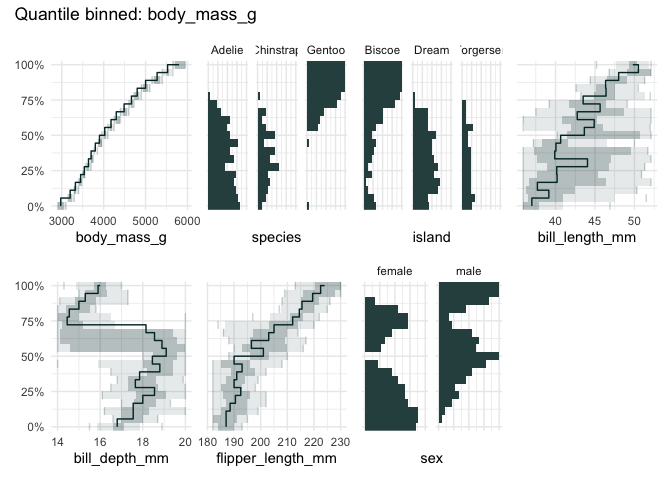
Or the well-known diamonds dataset
data("diamonds", package = "ggplot2")
table_plot(diamonds[1:7], "carat")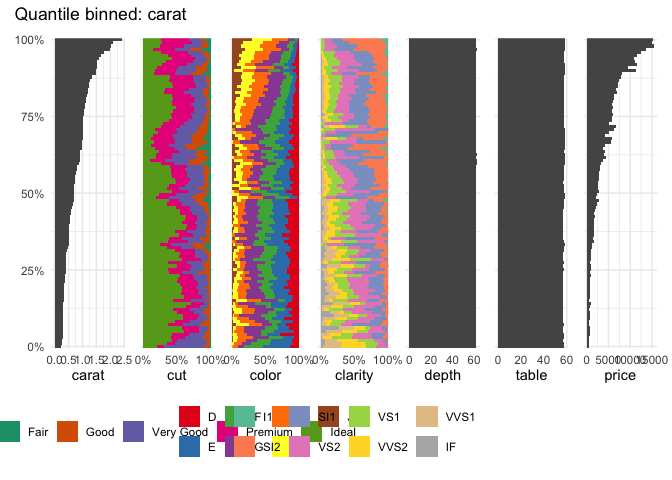
data("diamonds", package = "ggplot2")
qbin_boxplot(diamonds[1:7], "carat")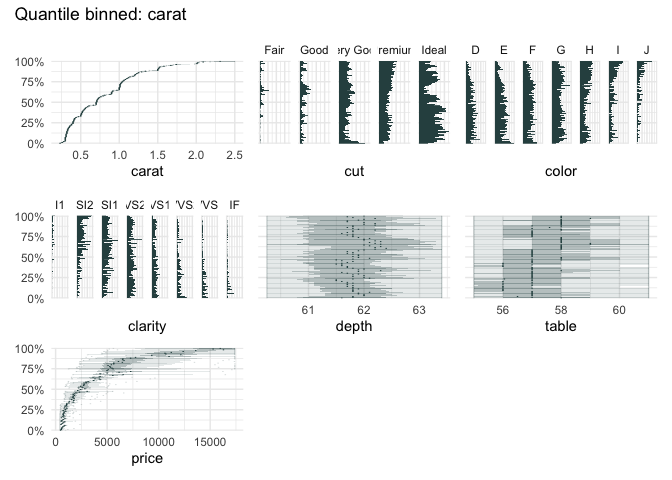
funq_plot(diamonds[1:7], "carat")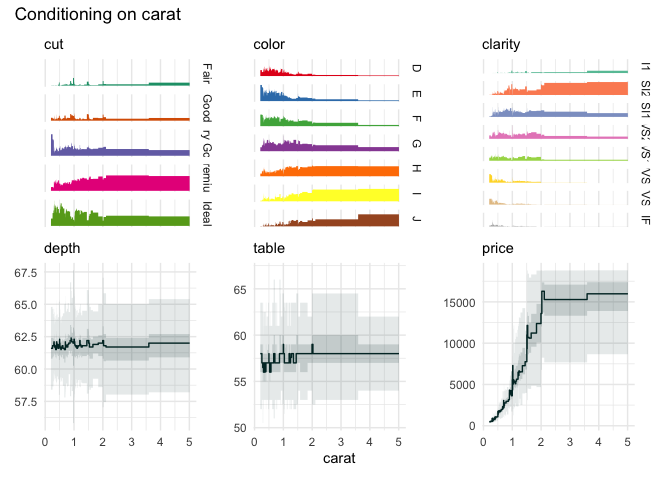
We can zoom in on the carat variable, because the upper
quantile bins are not very informative.
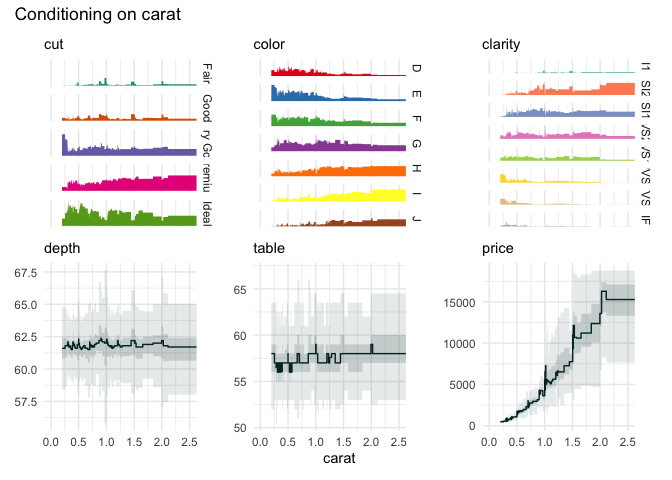
funq_plot(
diamonds[1:7],
"carat",
auto_fill = TRUE,
xlim = c(0, 2.5)
)
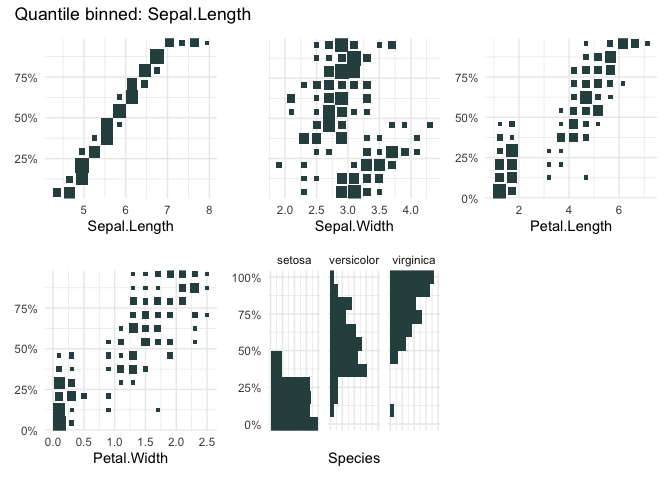
qbin_heatmap(
iris,
x = "Sepal.Length",
n = 12,
type="s"
)
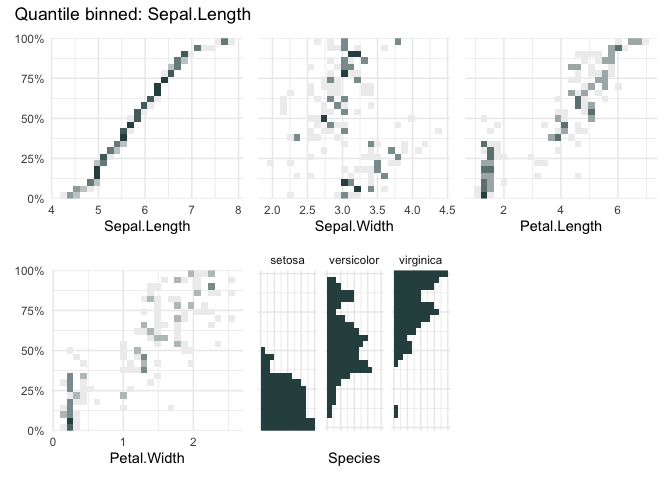
qbin_heatmap(
iris,
x = "Sepal.Length",
overlap = TRUE
)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.