The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The goal is to offer a package that can produce bias-corrected
performance measures for clinical prediction models with binary outcomes
for a range of model development approaches available in R (similar to
rms::validate). There are also functions for assessing
prediction stability, as described in Riley and Collins
(2023).
To install:
install.packages("pminternal") # cran
# or
devtools::install_github("stephenrho/pminternal") # developmentIn the example below we use bootstrapping to correct performance
measures for a glm via calculation of ‘optimism’ (see
vignette("pminternal"),
vignette("validate-examples"), and
vignette("missing-data") for more examples):
library(pminternal)
# make some data
set.seed(2345)
n <- 800
p <- 10
X <- matrix(rnorm(n*p), nrow = n, ncol = p)
LP <- -1 + apply(X[, 1:5], 1, sum) # first 5 variables predict outcome
y <- rbinom(n, 1, plogis(LP))
dat <- data.frame(y, X)
# fit a model
mod <- glm(y ~ ., data = dat, family = "binomial")
# calculate bootstrap optimism corrected performance measures
(val <- validate(fit = mod, method = "boot_optimism", B = 100))
#> It is recommended that B >= 200 for bootstrap validation
#> apparent optimism corrected n
#> C 0.8567 0.0093 0.8474 100
#> Brier 0.1423 -0.0054 0.1477 100
#> Intercept 0.0000 0.0175 -0.0175 100
#> Slope 1.0000 0.0529 0.9471 100
#> Eavg 0.0045 -0.0048 0.0093 100
#> E50 0.0039 -0.0050 0.0089 100
#> E90 0.0081 -0.0107 0.0187 100
#> Emax 0.0109 -0.0057 0.0165 100
#> ECI 0.0027 -0.0038 0.0065 100The other available methods for calculating bias corrected
performance are the simple bootstrap (boot_simple), 0.632
bootstrap optimism (.632), optimism via cross-validation
(cv_optimism), and regular cross-validation
(cv_average). Please see ?pminternal::validate
and the references therein. Bias corrected calibration curves can also
be produced (see pminternal::cal_plot). Confidence
intervals can also be added via confint.
For models that cannot be supported via fit, users are
able to specify their own model (model_fun) and prediction
(pred_fun) functions as shown below. Note that when
specifying user-defined model and prediction functions the data and
outcome must also be provided. It is crucial that model_fun
implements the entire model development procedure (variable selection,
hyperparameter tuning, etc). For more examples, see
vignette("pminternal") and
vignette("validate-examples").
# fit a glm with lasso penalty
library(glmnet)
#> Loading required package: Matrix
#> Loaded glmnet 4.1-8
lasso_fun <- function(data, ...){
y <- data$y
x <- as.matrix(data[, which(colnames(data) != "y")])
cv <- cv.glmnet(x=x, y=y, alpha=1, nfolds = 10, family="binomial")
lambda <- cv$lambda.min
glmnet(x=x, y=y, alpha = 1, lambda = lambda, family="binomial")
}
lasso_predict <- function(model, data, ...){
y <- data$y
x <- as.matrix(data[, which(colnames(data) != "y")])
predict(model, newx = x, type = "response")[,1]
}
(val <- validate(data = dat, outcome = "y",
model_fun = lasso_fun, pred_fun = lasso_predict,
method = "boot_optimism", B = 100))
#> It is recommended that B >= 200 for bootstrap validation
#> apparent optimism corrected n
#> C 0.856 0.0070 0.849 100
#> Brier 0.143 -0.0041 0.147 100
#> Intercept 0.080 0.0191 0.061 100
#> Slope 1.155 0.0449 1.110 100
#> Eavg 0.020 0.0013 0.019 100
#> E50 0.019 0.0026 0.017 100
#> E90 0.040 0.0021 0.038 100
#> Emax 0.044 0.0145 0.029 100
#> ECI 0.053 0.0087 0.044 100The output of validate (with
method = "boot_*") can be used to produce plots for
assessing the stability of model predictions (across models developed on
bootstrap resamples).
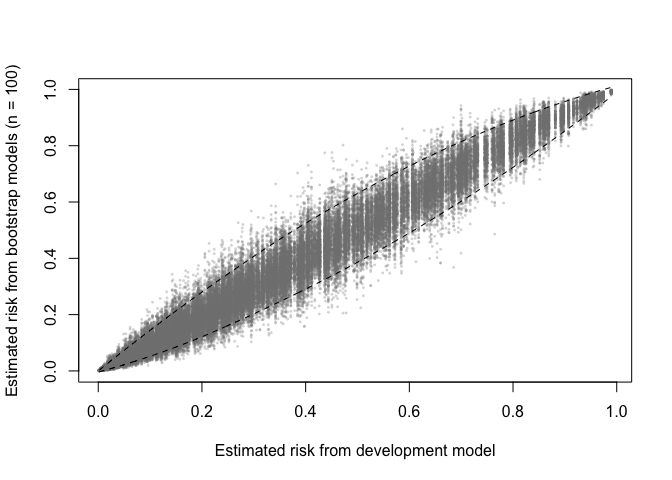
A prediction (in)stability plot shows predictions from the
B (in this case 100) bootstrap models applied to the
development data.
prediction_stability(val, smooth_bounds = TRUE)
A MAPE plot shows the mean absolute prediction error, which is the
difference between the predicted risk from the development model and
each of the B bootstrap models.
mape_stability(val)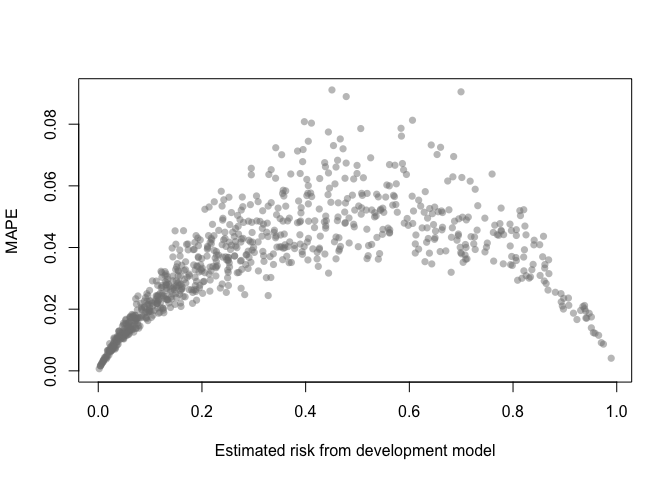
A calibration (in)stability plot depict the original calibration
curve along with B calibration curves from the bootstrap
models applied to the original data (y).
calibration_stability(val)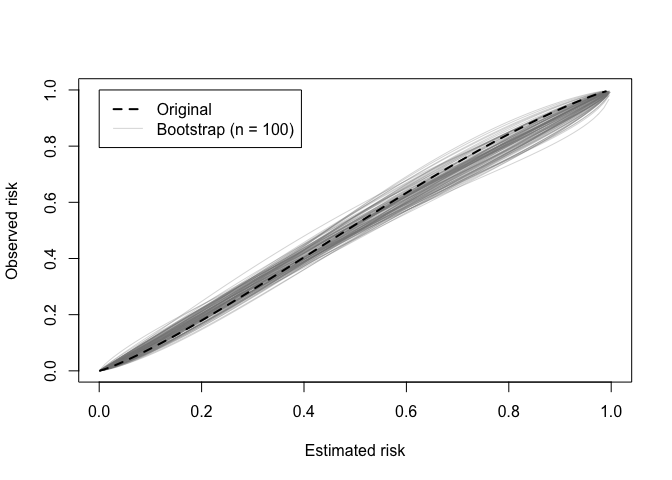
The classification instability index (CII) is the proportion of individuals that change predicted class (present/absent, 1/0) when predicted risk is compared to some threshold. For example, a patient predicted to be in class 1 would receive a CII of 0.3 if 30% of the bootstrap models led to a predicted class of 0.
classification_stability(val, threshold = .4)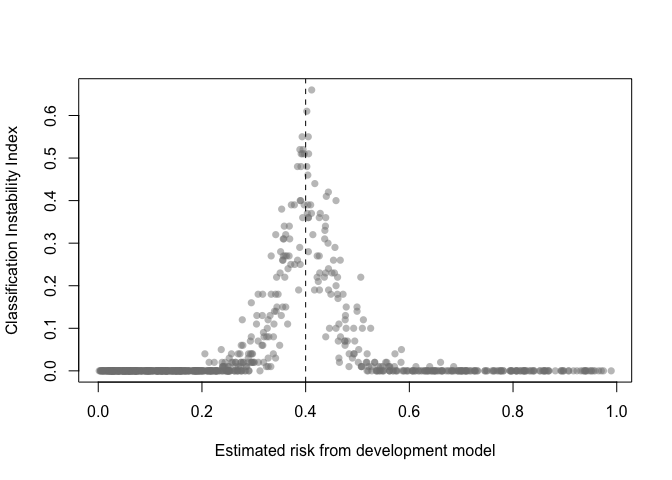
Decision curves implied by the original and bootstrap models can also be plotted.
dcurve_stability(val)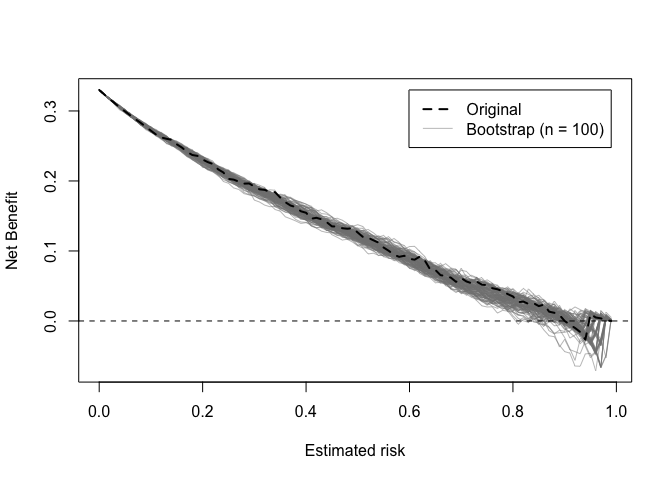
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.