The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

phylospatial is an R package for spatial phylogenetic
diversity analysis—accounting for evolutionary relationships among taxa
when describing biodiversity patterns. The package provides functions
for building and analyzing phylospatial data:
phylospatial() constructs a spatial phylogenetic data
set from community data and a tree.ps_diversity() calculates a range of phylogenetic
diversity and endemism metrics.ps_rand() computes significance values for diversity
metrics using null model randomizations.ps_dissim() calculates a pairwise community
phylogenetic beta diversity matrix.ps_ordinate() performs a community ordination to reduce
the dimensionality of the data set.ps_regions() clusters sites into phylogenetically
similar biogeographic regions.ps_prioritize() performs a spatial optimization to
identify conservation priorities.A key feature of phylospatial is full support for
quantitative community data, including occurrence probabilities (e.g.,
from species distribution models) and abundances, in addition to binary
presence-absence data. This avoids information loss from thresholding
continuous data, and enables new analyses not available in other spatial
phylogenetic R packages.
vignette("phylospatial-data") gives details about
constructing phylospatial datasets with different types of
data.vignette("alpha-diversity") demonstrates calculation of
alpha phylogenetic diversity and endemism measures, including
statistical hypothesis testing using randomization-based null
models.vignette("beta-diversity") shows how to calculate
phylogenetic beta diversity measures including nestedness and turnover,
as well as phylogenetic ordination and regionalization to visualize
phylogenetic community structure.vignette("prioritization") explains how to perform a
phylogenetic conservation prioritization.# you can install the package from CRAN:
install.packages("phylospatial")
# or the development version from GitHub:
remotes::install_github("matthewkling/phylospatial")library(phylospatial)
ps <- moss() # load example data
div <- ps_diversity(ps) # calculate diversity metrics
terra::plot(div)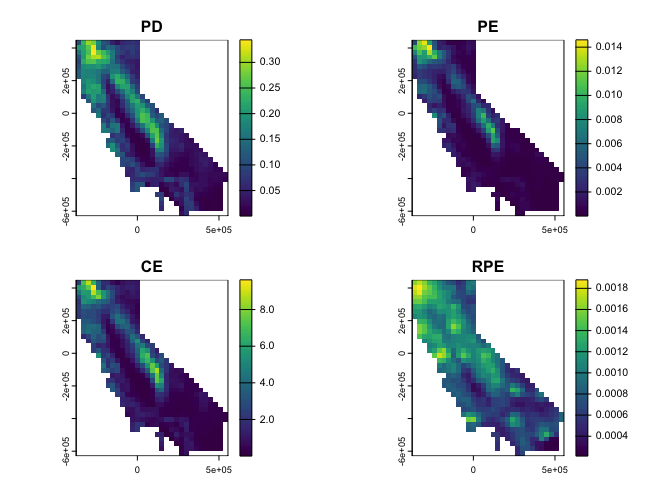
To cite phylospatial in publications, please use:
Kling, M. (2025). phylospatial: an R package for spatial phylogenetic analysis with quantitative community data. Methods in Ecology and Evolution 16:1075–1083. DOI 10.1111/2041-210x.70056
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.