
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

Taxonomic backbone and name validation tools for the mammals of Peru.
perumammals provides a curated, standardized and programmatically accessible version of the mammal diversity of Peru as compiled by Pacheco et al. (2021): “Lista actualizada de la diversidad de los mamíferos del Perú y una propuesta para su actualización”.
This publication represents the most up-to-date and comprehensive synthesis of Peruvian mammal diversity, integrating taxonomic revisions, biogeographic information, distributional updates and the evaluation of endemic taxa.
The package includes:
The goal of the package is not to replace authoritative taxonomic databases, but to provide a stable, Peruvian-focused backbone and a set of reproducible tools that can be used in ecological, environmental, biogeographic and conservation workflows.
The backbone included in perumammals is derived directly from the annex of Pacheco et al. (2021), who synthesized decades of Peruvian mammalogy work. This list is highly relevant as it incorporates recent taxonomic updates up to November 2021, including the description of species new to science (e.g., Thomasomys antoniobracki, Oligoryzomys guille), the first Peruvian records for some bats (Eumops bonariensis), and species re-validations (e.g., Neacomys carceleni), ensuring users work with the most current classification.
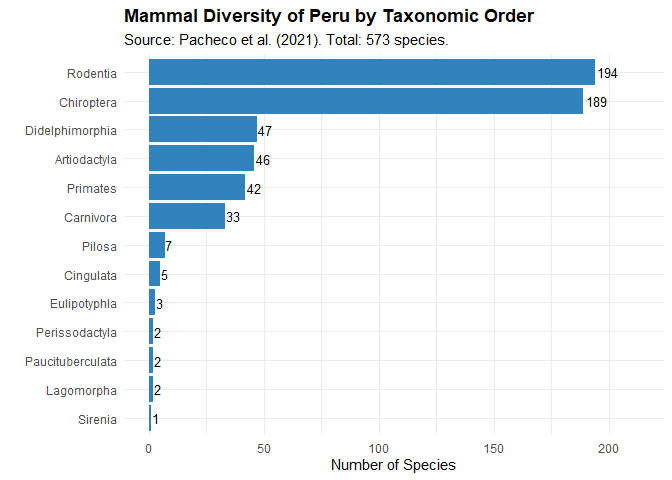

The article assigns each species to one or more Peruvian ecoregions using the classification widely used in biogeography and conservation planning:
#> ── Peruvian Mammal Ecoregions (Brack-Egg, 1986) ────────────────────────────────
#> ℹ Number of ecoregions: 10
#> ℹ Total mammal species in Peru: 573
#>
#> Ecoregions by species richness:
#>
#> SB - Selva Baja: 320 species (55.8%)
#> YUN - Yungas: 256 species (44.7%)
#> SP - Sabana de Palmera: 83 species (14.5%)
#> BSE - Bosque Seco Ecuatorial: 81 species (14.1%)
#> VOC - Vertiente Occidental: 72 species (12.6%)
#> PUN - Puna: 71 species (12.4%)
#> BPP - Bosque Pluvial del Pacífico: 69 species (12%)
#> COS - Costa: 66 species (11.5%)
#> OCE - Oceánica: 30 species (5.2%)
#> PAR - Páramo: 26 species (4.5%)
#>
#> Use pm_by_ecoregion() to filter species by ecoregion
#> Use include_endemic = TRUE to see endemic species counts
#> ────────────────────────────────────────────────────────────────────────────────These codes are incorporated into the package as both:
Pacheco et al. (2021) incorporate:
Although perumammals contains functions for name
validation and exploration.
The package includes four main datasets:
peru_mammalsA species-level backbone with:
peru_mammals_ecoregionsA long-format dataset listing species–ecoregion pairs, ideal for:
peru_mammals_ecoregions_metaMetadata describing the ecoregion codes used across the package.
peru_mammals_backboneMetadata describing:
These datasets make perumammals a lightweight but
powerful reference for any workflow requiring curated and
Peruvian-focused mammal information.
| Category | Functionality | Conceptual Code Example |
|---|---|---|
| Name Validation | Validate species names against database | validate_peru_mammals(c("Thomasomys notatus", "Tapirus terrestris", "Unknown species")) |
| Quick Checks | Check if species occurs in Peru | is_peru_mammal("Tremarctos ornatus") |
| Endemism Query | Check endemic status | is_endemic_peru("Thomasomys notatus") |
| Match Quality | Get validation match level | match_quality_peru("Puma concolar") |
| Family Summary | List families with species counts | pm_list_families() |
| Family Filter | Filter by specific family | pm_species(family = "Cricetidae") |
| Endemic Analysis | List endemic species statistics | pm_list_endemic() |
| Endemic by Family | Filter endemics by family | pm_endemics(family = "Phyllostomidae") |
| Endemic by Ecoregion | Filter endemics by ecoregion | pm_by_ecoregion(ecoregion = "YUN", endemic = TRUE) |
## Install from CRAN (recommended)
pak::pak("perumammals")
# Development version from GitHub
# Using pak (recommended)
pak::pak("PaulESantos/perumammals")
# Or using remotes
remotes::install_github("PaulESantos/perumammals")library(perumammals)
── Attaching perumammals ───────────────────────────────────────────────────────────────── perumammals 0.0.0.1 ──
✔ Taxonomic backbone: Pacheco et al. (2021) | Species: 573
ℹ Use pm_backbone_info() for full citation and detailsIf you use this package, please cite:
The package:
citation("perumammals")
#> To cite perumammals in publications, please use:
#>
#> Santos Andrade, P. E., & Gonzales Guillen, F. N. (2025). perumammals:
#> Taxonomic Backbone and Name Validation Tools for Mammals of Peru. R
#> package version 0.0.0.1. https://paulesantos.github.io/perumammals/
#>
#> The taxonomic backbone included in this package is based on:
#>
#> Pacheco, V., Cadenillas, R., Zeballos, H., Hurtado, C. M., Ruelas, D.,
#> & Pari, A. (2021). Lista actualizada de la diversidad de los mamíferos
#> del Perú y una propuesta para su actualización. Revista Peruana de
#> Biología, 28(special issue), e21019.
#> https://doi.org/10.15381/rpb.v28i4.21019
#>
#> To see these entries in BibTeX format, use 'print(<citation>,
#> bibtex=TRUE)', 'toBibtex(.)', or set
#> 'options(citation.bibtex.max=999)'.These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.