
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

Visualize simple 2-D decision tree partitions in R. The parttree package provides visualization methods for both base R graphics (via tinyplot) and ggplot2.
The stable version of parttree is available on CRAN.
install.packages("parttree")Or, you can grab the latest development version from R-universe.
install.packages("parttree", repos = "https://grantmcdermott.r-universe.dev")The parttree homepage includes an introductory vignette and detailed documentation. But here’s a quickstart example using the “kyphosis” dataset that comes bundled with the rpart package. In this case, we are interested in predicting kyphosis recovery after spinal surgery, as a function of 1) the number of topmost vertebra that were operated, and 2) patient age.
The key function is parttree(), which comes with its own
plotting method.
library(rpart) # For the dataset and fitting decisions trees
library(parttree) # This package
fit = rpart(Kyphosis ~ Start + Age, data = kyphosis)
# Grab the partitions and plot
fit_pt = parttree(fit)
plot(fit_pt)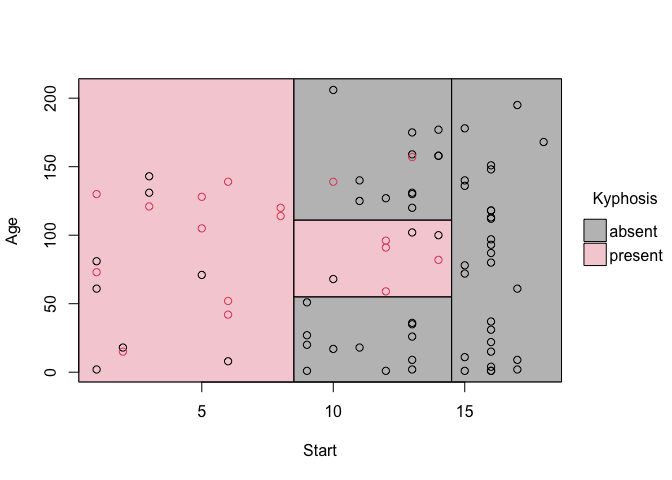
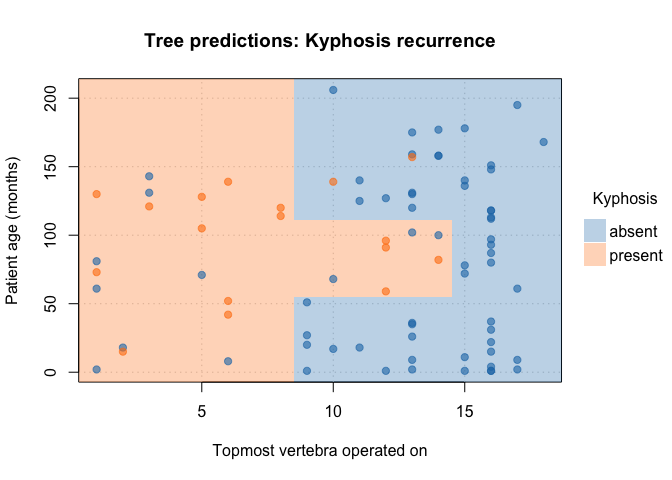
Customize your plots by passing additional arguments:
plot(
fit_pt,
border = NA, # no partition borders
pch = 19, # filled points
alpha = 0.6, # point transparency
grid = TRUE, # background grid
palette = "classic", # new colour palette
xlab = "Topmost vertebra operated on", # custom x title
ylab = "Patient age (months)", # custom y title
main = "Tree predictions: Kyphosis recurrence" # custom title
)
For ggplot2 users, we offer an equivalent workflow
via the geom_partree() visualization layer.
library(ggplot2) ## Should be loaded separately
ggplot(kyphosis, aes(x = Start, y = Age)) +
geom_parttree(data = fit, alpha = 0.1, aes(fill = Kyphosis)) + # <-- key layer
geom_point(aes(col = Kyphosis)) +
labs(
x = "No. of topmost vertebra operated on", y = "Patient age (months)",
caption = "Note: Points denote observations. Shading denotes model predictions."
) +
theme_minimal()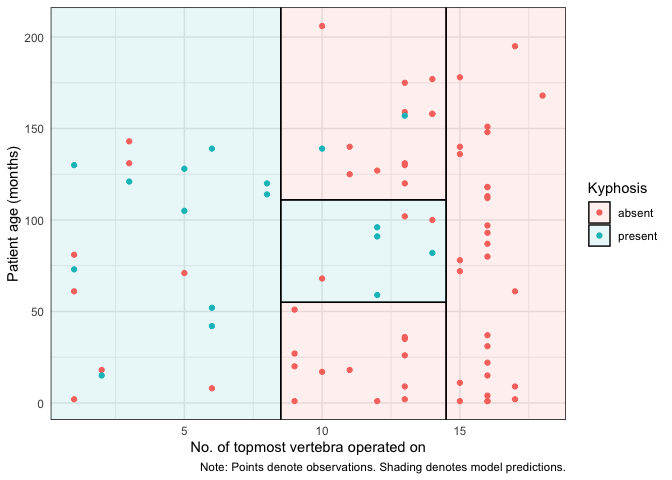
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.