
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

| Project | Main | Devel |
|---|---|---|
 |
||
mpmsim contains tools for generating random or
semi-random matrix population models (MPMs) given a particular life
history archetype. It also facilitates the generation of Leslie
matrices, and the simulation of MPMs based on expected transition rates
and sample sizes. This can be useful for exploring uncertainty in
inferences when sample sizes are small (or unknown).
You can install the latest stable version of mpmsim from
CRAN like this:
install.packages("mpmsim")The package is being developed (here) on GitHub. You can install the
latest development version of mpmsim like this:
# install package 'remotes' if necessary
# will already be installed if 'devtools' is installed
install.packages("remotes")
# argument 'build_opts = NULL' only needed if you want to build vignettes
remotes::install_github("jonesor/mpmsim", build_opts = NULL)During development there may be other versions, with additional functionality, available on different GitHub “branches”. To install from one of these branches, use the following syntax:
# install from the 'dev' branch
remotes::install_github("jonesor/mpmsim", ref = "dev")First, load the package.
library(mpmsim)The make_leslie_mpm function can be used to generate a
Leslie matrix model (Leslie, 1945) where the stages represent discrete
age classes (usually years of life).
In a Leslie matrix, survival is represented in the lower sub-diagonal and the lower-right-hand corner element, while fecundity (reproduction) is shown in the top row. Both survival and fecundity have a length equal to the number of stages in the model. Users can specify both survival and fecundity as either a single value or a vector of values, with a length equal to the dimensions of the matrix model. If these arguments are single values, the value is repeated along the survival/fecundity sequence.
make_leslie_mpm(
survival = seq(0.1, 0.45, length.out = 4),
fecundity = c(0, 0, 2.4, 5), n_stages = 4, split = FALSE
)
#> [,1] [,2] [,3] [,4]
#> [1,] 0.0 0.0000000 2.4000000 5.00
#> [2,] 0.1 0.0000000 0.0000000 0.00
#> [3,] 0.0 0.2166667 0.0000000 0.00
#> [4,] 0.0 0.0000000 0.3333333 0.45Users can generate Leslie matrices with particular functional forms
of mortality by first making a data frame of a simplified life table
that includes age and survival probability within each age interval. The
model_mortality function can handle the following models:
Gompertz, Gompertz-Makeham, Weibull, Weibull-Makeham, Siler and
Exponential.
The function returns a standard life table data.frame
including columns for age (x), age-specific hazard
(hx), survivorship (lx), age-specific
probability of death and survival (qx and px).
By default, the life table is truncated at the age when the survivorship
function declines below 0.01 (i.e. when only 1% of individuals in a
cohort would remain alive).
For example to produce a life table based on Gompertz mortality:
(surv_prob <- model_mortality(params = c(0.2, 0.4), model = "Gompertz"))
#> x hx lx qx px
#> 1 0 0.2000000 1.00000000 0.2205623 0.7794377
#> 2 1 0.2983649 0.77943774 0.3104641 0.6895359
#> 3 2 0.4451082 0.53745028 0.4256784 0.5743216
#> 4 3 0.6640234 0.30866930 0.5627783 0.4372217
#> 5 4 0.9906065 0.13495691 0.7089351 0.2910649
#> 6 5 1.4778112 0.03928123 0.8413767 0.1586233Users can also use a functional form for fecundity (see
?model_fecundity), including, logistic, step, von
Bertalanffy, Normal and Hadwiger.
Here a simple step function is assumed.
survival <- surv_prob$px
fecundity <- model_fecundity(
age = 0:(length(survival) - 1),
params = c(A = 5), maturity = 2, model = "step"
)Subsequently, these survival and fecundity values can be applied to the Leslie matrix as follows.
make_leslie_mpm(
survival = survival, fecundity = fecundity,
n_stages = length(survival), split = FALSE
)
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 0.0000000 0.0000000 5.0000000 5.0000000 5.0000000 5.0000000
#> [2,] 0.7794377 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.0000000 0.6895359 0.0000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.0000000 0.0000000 0.5743216 0.0000000 0.0000000 0.0000000
#> [5,] 0.0000000 0.0000000 0.0000000 0.4372217 0.0000000 0.0000000
#> [6,] 0.0000000 0.0000000 0.0000000 0.0000000 0.2910649 0.1586233Users can generate large numbers of plausible Leslie matrices using
the rand_leslie_set function.
The arguments for this function include the number of models
(n_models), the type of mortality
(e.g. GompertzMakeham) and reproduction
(e.g. step). The specific parameters for mortality and
reproduction are provided as defined distributions from which parameters
can be drawn at random. The type of distribution is defined with the
dist_type argument and can be uniform or
normal, and the distributions are defined using the
mortality_params and fecundity_params
arguments, which accept data frames of distribution parameters.
For example, the following code produces a list of five Leslie matrices that have Gompertz-Makeham mortality characteristics and where reproduction is a step function.
First, we define the limits of a uniform distributions for the Gompertz mortality and step fecundity functions.
mortParams <- data.frame(
minVal = c(0.05, 0.08, 0.7),
maxVal = c(0.14, 0.15, 0.7)
)
fecParams <- data.frame(minVal = 4, maxVal = 6)We also set maturity to be drawn from a distribution ranging from 0 to 3.
maturityParams <- c(0, 3)Now we produce the models. We output as “Type5” which is
a simple list of the main A matrix model, but outputs can also be split
into submatrices (e.g. the U and F matrices), or as a
CompadreDB object.
outputMPMs <- rand_leslie_set(
n_models = 5, mortality_model = "GompertzMakeham", fecundity_model = "step",
mortality_params = mortParams,
fecundity_params = fecParams,
fecundity_maturity_params = maturityParams,
dist_type = "uniform",
output = "Type5"
)
outputMPMs
#> [[1]]
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 0.0000000 0.0000000 0.0000000 4.5722791 4.5722791 4.5722791
#> [2,] 0.4305453 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.0000000 0.4210229 0.0000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.0000000 0.0000000 0.4102704 0.0000000 0.0000000 0.0000000
#> [5,] 0.0000000 0.0000000 0.0000000 0.3981747 0.0000000 0.0000000
#> [6,] 0.0000000 0.0000000 0.0000000 0.0000000 0.3846275 0.3695309
#>
#> [[2]]
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 0.0000000 5.4731766 5.473177 5.4731766 5.4731766 5.4731766
#> [2,] 0.4429031 0.0000000 0.000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.0000000 0.4366956 0.000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.0000000 0.0000000 0.429826 0.0000000 0.0000000 0.0000000
#> [5,] 0.0000000 0.0000000 0.000000 0.4222377 0.0000000 0.0000000
#> [6,] 0.0000000 0.0000000 0.000000 0.0000000 0.4138729 0.4046735
#>
#> [[3]]
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 0.0000000 0.000000 0.0000000 4.9154836 4.9154836 4.9154836
#> [2,] 0.4419032 0.000000 0.0000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.0000000 0.434841 0.0000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.0000000 0.000000 0.4269406 0.0000000 0.0000000 0.0000000
#> [5,] 0.0000000 0.000000 0.0000000 0.4181238 0.0000000 0.0000000
#> [6,] 0.0000000 0.000000 0.0000000 0.0000000 0.4083108 0.3974225
#>
#> [[4]]
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 0.000000 0.0000000 0.0000000 4.9245856 4.9245856 4.9245856
#> [2,] 0.431272 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.000000 0.4250633 0.0000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.000000 0.0000000 0.4183198 0.0000000 0.0000000 0.0000000
#> [5,] 0.000000 0.0000000 0.0000000 0.4110069 0.0000000 0.0000000
#> [6,] 0.000000 0.0000000 0.0000000 0.0000000 0.4030901 0.3945359
#>
#> [[5]]
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 0.0000000 0.0000000 4.9499942 4.9499942 4.9499942 4.9499942
#> [2,] 0.4298125 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.0000000 0.4241257 0.0000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.0000000 0.0000000 0.4180004 0.0000000 0.0000000 0.0000000
#> [5,] 0.0000000 0.0000000 0.0000000 0.4114112 0.0000000 0.0000000
#> [6,] 0.0000000 0.0000000 0.0000000 0.0000000 0.4043329 0.3967408The rand_lefko_mpm function can be used to generate a
random Lefkovitch matrix population model (MPM) (Lefkovitch, 1965), with
element values based on defined life history archetypes.
The function draws survival and transition/growth probabilities from
a Dirichlet distribution to ensure that the column totals, including
death, are less than or equal to 1. Fecundity can be specified as a
single value or as a vector with a length equal to the dimensions of the
matrix. If specified as a single value, it is placed in the top-right
corner of the matrix. If specified as a vector of length
n_stages, it spans the entire top row of the matrix. The
archetype argument can be used to constrain the MPMs, for
example, archetype = 2 constraints the survival probability
to increase monotonically as individuals advance to later stages.
For more information, see the documentation for
rand_lefko_mpm and Takada et al. (2018), from which these
archetypes are derived.
In the following example, I split the output matrices into the
U and F submatrices, which can be summed to
create the full A matrix model.
(rMPM <- rand_lefko_mpm(
n_stages = 3, fecundity = 20,
archetype = 2, split = TRUE
))
#> $mat_A
#> [,1] [,2] [,3]
#> [1,] 0.2070973 0.33155927 20.4132432
#> [2,] 0.3836494 0.52219726 0.3625132
#> [3,] 0.2615892 0.03314957 0.1157180
#>
#> $mat_U
#> [,1] [,2] [,3]
#> [1,] 0.2070973 0.33155927 0.4132432
#> [2,] 0.3836494 0.52219726 0.3625132
#> [3,] 0.2615892 0.03314957 0.1157180
#>
#> $mat_F
#> [,1] [,2] [,3]
#> [1,] 0 0 20
#> [2,] 0 0 0
#> [3,] 0 0 0The rand_lefko_set function can be used to quickly
generate large numbers of Lefkovitch MPMs using the above approach. For
example, the following code generates five MPMs with archetype 1. By
using the constraint argument, users can specify an
acceptable characteristics for the set of matrices. In this case,
population growth rate range, which can be useful for life history
analyses where we might assume that only life histories with lambda
values close to 1 can persist in nature. We set the argument
output = "Type5" to ensure that the function returns a
list object.
library(popbio)
constrain_df <- data.frame(fun = "lambda", arg = NA, lower = 0.9, upper = 1.1)
rand_lefko_set(
n_models = 5, n_stages = 4, fecundity = 8, archetype = 1, constraint = constrain_df,
output = "Type5"
)
#> [[1]]
#> [,1] [,2] [,3] [,4]
#> [1,] 0.28730926 0.02716436 0.26331722 8.46373314
#> [2,] 0.14460260 0.15628773 0.23535192 0.02222792
#> [3,] 0.10395162 0.24279393 0.10570287 0.17071769
#> [4,] 0.03134086 0.27716832 0.01175425 0.07549711
#>
#> [[2]]
#> [,1] [,2] [,3] [,4]
#> [1,] 0.14077752 0.04357884 0.429128096 8.0837046
#> [2,] 0.09905905 0.52812214 0.007308617 0.2701657
#> [3,] 0.36955076 0.11374572 0.109339485 0.2160414
#> [4,] 0.01698186 0.01869725 0.143428706 0.1214954
#>
#> [[3]]
#> [,1] [,2] [,3] [,4]
#> [1,] 0.160744755 0.02845733 0.03688629 8.1365669
#> [2,] 0.041433197 0.24550232 0.01277293 0.1219770
#> [3,] 0.791265908 0.02813589 0.25420572 0.2599794
#> [4,] 0.002908193 0.21314599 0.04493534 0.3332529
#>
#> [[4]]
#> [,1] [,2] [,3] [,4]
#> [1,] 0.196022839 0.39576976 0.27489845 8.24843871
#> [2,] 0.350613432 0.10892595 0.28872665 0.05133337
#> [3,] 0.084225194 0.23979127 0.19811975 0.41727119
#> [4,] 0.009066956 0.06365681 0.09946455 0.11853471
#>
#> [[5]]
#> [,1] [,2] [,3] [,4]
#> [1,] 0.04407168 0.09512729 0.03927867 8.00717018
#> [2,] 0.07098214 0.28541167 0.23663234 0.49786947
#> [3,] 0.36675467 0.46916408 0.06540892 0.13898581
#> [4,] 0.05606256 0.11920243 0.03335758 0.08196272Sometimes, users may find themselves confronted with an MPM for which
they can calculate various metrics, and have a need to calculate the
confidence interval for those metrics. The compute_ci
function is designed to address this need by computing 95% confidence
intervals (CIs) for measures derived from a complete MPM (i.e. the A
matrix).
This is accomplished using parametric bootstrapping, generating a sampling distribution of the MPM by performing numerous random independent draws using the sampling distribution of each underlying transition rate. The approach relies on (1) a known (or estimated) sample size for each estimate in the model and (2) the assumption that survival-related processes are binomial, while reproduction processes follow a Poisson distribution.
Here’s an example, where we use the Lefkovitch model from above, and where we believe the sample size was 10 individuals for each parameter estimate.
The point estimate for population growth rate (lambda) is 2.539.
library(popdemo)
eigs(rMPM$mat_A, what = "lambda")
#> [1] 2.539016Users can calculate the 95% CI, assuming a sample size of 10, like this:
compute_ci(
mat_U = rMPM$mat_U, mat_F = rMPM$mat_F,
sample_size = 10,
FUN = eigs, what = "lambda"
)
#> 2.5% 97.5%
#> 0.8384508 3.4177693The sample_size argument can handle various cases, for
example, where sample size varies across the matrix, or between the U
and F submatrices (see ?compute_ci).
An equivalent function, compute_ci_U is designed for use
when the derived estimate requires only the U submatrix (as opposed to
both submatrices of the A matrix).
The function add_mpm_error can be used to simulate an
MPM with sampling error, based on expected transition rates (survival
and fecundity) and sample sizes. This could be useful at the initial
phases of a study, as part of a power analysis, or could be used simply
to get a feel for expected variation under different circumstances.
The expected transition rates must be provided as matrices. The sample size(s) can be given as either a matrix of sample sizes for each element of the matrix or as a single value which is then applied to all elements of the matrix.
The function uses a binomial process to simulate survival/growth elements and a Poisson process to simulate the fecundity elements. As a result, when sample sizes are large, the simulated MPM will closely reflect the expected transition rates. In contrast, when sample sizes are small, the simulated matrices will become more variable.
To illustrate use of the function, the following code first generates
a 3-stage Leslie matrix using the make_leslie_mpm function.
It then passes the U and F matrices from this Leslie matrix to the
add_mpm_error function. Then, two matrices are simulated,
first with a sample size of 1000, and then with a sample size of
seven.
mats <- make_leslie_mpm(
survival = c(0.3, 0.5, 0.8),
fecundity = c(0, 2.2, 4.4),
n_stages = 3, split = TRUE
)
add_mpm_error(
mat_U = mats$mat_U, mat_F = mats$mat_F,
sample_size = 1000, split = FALSE, by_type = FALSE
)
#> [,1] [,2] [,3]
#> [1,] 0.000 2.220 4.316
#> [2,] 0.293 0.000 0.000
#> [3,] 0.000 0.485 0.816
add_mpm_error(
mat_U = mats$mat_U, mat_F = mats$mat_F,
sample_size = 7, split = FALSE, by_type = FALSE
)
#> [,1] [,2] [,3]
#> [1,] 0.0000000 3.5714286 4.2857143
#> [2,] 0.1428571 0.0000000 0.0000000
#> [3,] 0.0000000 0.2857143 0.8571429A list of an arbitrary number of matrices can be generated easily
using replicate, as follows.
replicate(
n = 5,
add_mpm_error(
mat_U = mats$mat_U, mat_F = mats$mat_F,
sample_size = 7, split = FALSE, by_type = FALSE
)
)
#> , , 1
#>
#> [,1] [,2] [,3]
#> [1,] 0.0000000 1.5714286 4.7142857
#> [2,] 0.5714286 0.0000000 0.0000000
#> [3,] 0.0000000 0.2857143 0.8571429
#>
#> , , 2
#>
#> [,1] [,2] [,3]
#> [1,] 0.0000000 1.0000000 4.857143
#> [2,] 0.1428571 0.0000000 0.000000
#> [3,] 0.0000000 0.2857143 1.000000
#>
#> , , 3
#>
#> [,1] [,2] [,3]
#> [1,] 0.0000000 1.8571429 4.571429
#> [2,] 0.1428571 0.0000000 0.000000
#> [3,] 0.0000000 0.4285714 1.000000
#>
#> , , 4
#>
#> [,1] [,2] [,3]
#> [1,] 0.0000000 2.7142857 4.2857143
#> [2,] 0.4285714 0.0000000 0.0000000
#> [3,] 0.0000000 0.7142857 0.8571429
#>
#> , , 5
#>
#> [,1] [,2] [,3]
#> [1,] 0.0000000 2.0000000 4.4285714
#> [2,] 0.2857143 0.0000000 0.0000000
#> [3,] 0.0000000 0.8571429 0.7142857This could be coerced into a CompadreDB object, if
necessary, using the cdb_build_cdb function from the
Rcompadre package.
It can be helpful to visualise the matrices. This can be accomplished
with the function plot_matrix. The output of
plot_matrix is of class ggplot and as such the
colour scheme can be modified in the usual way with, for example,
scale_fill_gradient or similar.
Here’s the matrix:
rMPM$mat_U
#> [,1] [,2] [,3]
#> [1,] 0.2070973 0.33155927 0.4132432
#> [2,] 0.3836494 0.52219726 0.3625132
#> [3,] 0.2615892 0.03314957 0.1157180And here’s the plot:
p <- plot_matrix(rMPM$mat_U)
p + ggplot2::scale_fill_gradient(low = "black", high = "yellow")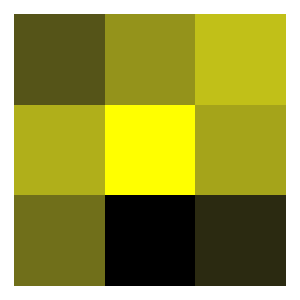
All contributions are welcome. Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
There are numerous ways of contributing.
You can submit bug reports, suggestions etc. by opening an issue.
You can copy or fork the repository, make your own code edits and then send us a pull request. Here’s how to do that.
You are also welcome to email me.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.