
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

Julia GLM.jl and MixedModels.jl based implementation of the cluster-based permutation test for time series data, powered by JuliaConnectoR.

Install the released version of jlmerclusterperm from CRAN:
install.packages("jlmerclusterperm")Or install the development version from GitHub with:
# install.packages("remotes")
remotes::install_github("yjunechoe/jlmerclusterperm")Using jlmerclusterperm requires a prior installation of
the Julia programming language, which can be downloaded from either the
official website or using the
command line utility juliaup. Julia version
>=1.8 is required and 1.9
or higher is preferred for the substantial speed improvements.
Before using functions from jlmerclusterperm, an initial
setup is required via calling jlmerclusterperm_setup(). The
very first call on a system will install necessary dependencies (this
only happens once and takes around 10-15 minutes).
Subsequent calls to jlmerclusterperm_setup() incur a
small overhead of around 30 seconds, plus slight delays for first-time
function calls. You pay up front for start-up and warm-up costs and get
blazingly-fast functions from the package.
# Both lines must be run at the start of each new session
library(jlmerclusterperm)
jlmerclusterperm_setup()
See the Get Started page on the package website for background and tutorials.
clusterpermute()A time series data:
chickweights <- ChickWeight
chickweights$Time <- as.integer(factor(chickweights$Time))
matplot(
tapply(chickweights$weight, chickweights[c("Time", "Diet")], mean),
type = "b", lwd = 3, ylab = "Weight", xlab = "Time"
)
Preparing a specification object with
make_jlmer_spec():
chickweights_spec <- make_jlmer_spec(
formula = weight ~ 1 + Diet,
data = chickweights,
subject = "Chick", time = "Time"
)
chickweights_spec
Cluster-based permutation test with
clusterpermute():
set_rng_state(123L)
clusterpermute(
chickweights_spec,
threshold = 2.5,
nsim = 100
)
Including random effects:
chickweights_re_spec <- make_jlmer_spec(
formula = weight ~ 1 + Diet + (1 | Chick),
data = chickweights,
subject = "Chick", time = "Time"
)
set_rng_state(123L)
clusterpermute(
chickweights_re_spec,
threshold = 2.5,
nsim = 100
)$empirical_clusters
Computing time-wise statistics of the observed data:
empirical_statistics <- compute_timewise_statistics(chickweights_spec)
matplot(t(empirical_statistics), type = "b", pch = 1, lwd = 3, ylab = "t-statistic")
abline(h = 2.5, lty = 3)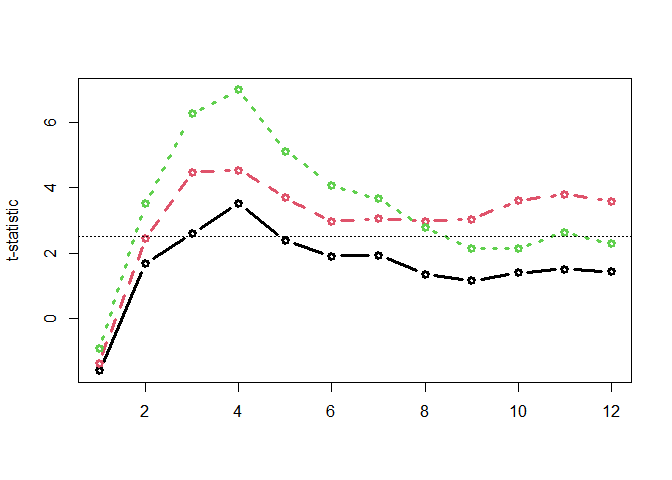
Identifying empirical clusters:
empirical_clusters <- extract_empirical_clusters(empirical_statistics, threshold = 2.5)
empirical_clusters
Simulating the null distribution:
set_rng_state(123L)
null_statistics <- permute_timewise_statistics(chickweights_spec, nsim = 100)
null_cluster_dists <- extract_null_cluster_dists(null_statistics, threshold = 2.5)
null_cluster_dists
Significance testing the cluster-mass statistic:
calculate_clusters_pvalues(empirical_clusters, null_cluster_dists, add1 = TRUE)
Iterating over a range of threshold values:
walk_threshold_steps(empirical_statistics, null_statistics, steps = c(2, 2.5, 3))
The paper Maris & Oostenveld (2007) which originally proposed the cluster-based permutation analysis.
The JuliaConnectoR package for powering the R interface to Julia.
The Julia packages GLM.jl and MixedModels.jl for fast implementations of (mixed effects) regression models.
Existing implementations of CPA in R (permuco, permutes, etc.) whose designs inspired the CPA interface in jlmerclusterperm.
If you use jlmerclusterperm for cluster-based permutation test with mixed-effects models in your research, please cite one (or more) of the following as you see fit.
To cite jlmerclusterperm:
To cite the cluster-based permutation test:
To cite the Julia programming language:
To cite the GLM.jl and MixedModels.jl Julia libraries, consult their Zenodo pages:
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.