
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

Gain insight into your models!
When fitting any statistical model, there are many useful pieces of information that are simultaneously calculated and stored beyond coefficient estimates and general model fit statistics. Although there exist some generic functions to obtain model information and data, many package-specific modelling functions do not provide such methods to allow users to access such valuable information.
insight is an R-package that fills this important gap by providing a suite of functions to support almost any model (see a list of the many models supported below in the List of Supported Packages and Models section). The goal of insight, then, is to provide tools to provide easy, intuitive, and consistent access to information contained in model objects. These tools aid applied research in virtually any field who fit, diagnose, and present statistical models by streamlining access to every aspect of many model objects via consistent syntax and output.
The insight package is available on CRAN, while its latest development version is available on R-universe (from rOpenSci) or GitHub.
| Type | Source | Command |
|---|---|---|
| Release | CRAN | install.packages("insight") |
| Development | r-universe | install.packages("insight", repos = "https://easystats.r-universe.dev") |
| Development | GitHub | remotes::install_github("easystats/insight") |
Once you have downloaded the package, you can then load it using:
library("insight")Tip
Instead of
library(insight), uselibrary(easystats). This will make all features of the easystats-ecosystem available.To stay updated, use
easystats::install_latest().
Built with non-programmers in mind, insight offers a
broad toolbox for making model and data information easily accessible.
While insight offers many useful functions for working
with and understanding model objects (discussed below), we suggest users
start with model_info(), as this function provides a clean
and consistent overview of model objects (e.g., functional form of the
model, the model family, link function, number of observations,
variables included in the specification, etc.). With a clear
understanding of the model introduced, users are able to adapt other
functions for more nuanced exploration of and interaction with virtually
any model object.Please visit https://easystats.github.io/insight/ for
documentation.
The functions from insight address different components of a model. In an effort to avoid confusion about specific “targets” of each function, in this section we provide a short explanation of insight’s definitions of regression model components.
The dataset used to fit the model.
Values estimated or learned from data that capture the relationship between variables. In regression models, these are usually referred to as coefficients.
Any unique variable names that appear in a regression model, e.g.,
response variable, predictors or random effects. A “variable” only
relates to the unique occurence of a term, or the term name. For
instance, the expression x + poly(x, 2) has only the
variable x.
Terms themselves consist of variable and factor names separated by
operators, or involve arithmetic expressions. For instance, the
expression x + poly(x, 2) has one variable
x, but two terms x and
poly(x, 2).
Aren’t the predictors, terms and parameters the same thing?
In some cases, yes. But not in all cases. Find out more by clicking here to access the documentation.
The package revolves around two key prefixes: get_* and
find_*. The get_* prefix extracts
values (or data) associated with model-specific
objects (e.g., parameters or variables), while the find_*
prefix lists model-specific objects (e.g., priors or
predictors). These are powerful families of functions allowing for great
flexibility in use, whether at a high, descriptive level
(find_*) or narrower level of statistical inspection and
reporting (get_*).
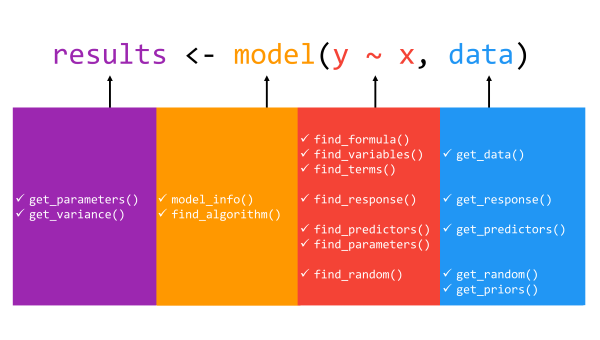
In total, the insight package includes 16 core
functions: get_data(),
get_priors(),
get_variance(),
get_parameters(),
get_predictors(),
get_random(),
get_response(),
find_algorithm(),
find_formula(),
find_variables(),
find_terms(),
find_parameters(),
find_predictors(),
find_random(),
find_response(),
and model_info().
In all cases, users must supply at a minimum, the name of the model fit
object. In several functions, there are additional arguments that allow
for more targeted returns of model information. For example, the
find_terms() function’s effects argument
allows for the extraction of “fixed effects” terms, “random effects”
terms, or by default, “all” terms in the model object. We point users to
the package documentation or the complementary package website, https://easystats.github.io/insight/, for a detailed
list of the arguments associated with each function as well as the
returned values from each function.
We now would like to provide examples of use cases of the insight package. These examples probably do not cover typical real-world problems, but serve as illustration of the core idea of this package: The unified interface to access model information. insight should help both users and package developers in order to reduce the hassle with the many exceptions from various modelling packages when accessing model information.
Say, the goal is to make predictions for a certain term, holding
remaining co-variates constant. This is achieved by calling
predict() and feeding the newdata-argument
with the values of the term of interest as well as the “constant” values
for remaining co-variates. The functions get_data() and
find_predictors() are used to get this information, which
then can be used in the call to predict().
In this example, we fit a simple linear model, but it could be replaced by (m)any other models, so this approach is “universal” and applies to many different model objects.
library(insight)
m <- lm(
Sepal.Length ~ Species + Petal.Width + Sepal.Width,
data = iris
)
dat <- get_data(m)
pred <- find_predictors(m, flatten = TRUE)
l <- lapply(pred, function(x) {
if (is.numeric(dat[[x]])) {
mean(dat[[x]])
} else {
unique(dat[[x]])
}
})
names(l) <- pred
l <- as.data.frame(l)
cbind(l, predictions = predict(m, newdata = l))
#> Species Petal.Width Sepal.Width predictions
#> 1 setosa 1.2 3.1 5.1
#> 2 versicolor 1.2 3.1 6.1
#> 3 virginica 1.2 3.1 6.3The next example should emphasize the possibilities to generalize functions to many different model objects using insight. The aim is simply to print coefficients in a complete, human readable sentence.
The first approach uses the functions that are available for some, but obviously not for all models, to access the information about model coefficients.
print_params <- function(model) {
paste0(
"My parameters are ",
toString(row.names(summary(model)$coefficients)),
", thank you for your attention!"
)
}
m1 <- lm(Sepal.Length ~ Petal.Width, data = iris)
print_params(m1)
#> [1] "My parameters are (Intercept), Petal.Width, thank you for your attention!"
# obviously, something is missing in the output
m2 <- mgcv::gam(Sepal.Length ~ Petal.Width + s(Petal.Length), data = iris)
print_params(m2)
#> [1] "My parameters are , thank you for your attention!"As we can see, the function fails for gam-models. As the access to models depends on the type of the model in the R ecosystem, we would need to create specific functions for all models types. With insight, users can write a function without having to worry about the model type.
print_params <- function(model) {
paste0(
"My parameters are ",
toString(insight::find_parameters(model, flatten = TRUE)),
", thank you for your attention!"
)
}
m1 <- lm(Sepal.Length ~ Petal.Width, data = iris)
print_params(m1)
#> [1] "My parameters are (Intercept), Petal.Width, thank you for your attention!"
m2 <- mgcv::gam(Sepal.Length ~ Petal.Width + s(Petal.Length), data = iris)
print_params(m2)
#> [1] "My parameters are (Intercept), Petal.Width, s(Petal.Length), thank you for your attention!"In case you want to file an issue or contribute in another way to the package, please follow this guide. For questions about the functionality, you may either contact us via email or also file an issue.
Currently, about 251 model classes are supported.
supported_models()
#> [1] "aareg" "afex_aov"
#> [3] "AKP" "Anova.mlm"
#> [5] "anova.rms" "aov"
#> [7] "aovlist" "Arima"
#> [9] "asym" "averaging"
#> [11] "bamlss" "bamlss.frame"
#> [13] "bayesQR" "bayesx"
#> [15] "BBmm" "BBreg"
#> [17] "bcplm" "betamfx"
#> [19] "betaor" "betareg"
#> [21] "BFBayesFactor" "bfsl"
#> [23] "BGGM" "bife"
#> [25] "bifeAPEs" "bigglm"
#> [27] "biglm" "blavaan"
#> [29] "blrm" "bracl"
#> [31] "brglm" "brmsfit"
#> [33] "brmultinom" "btergm"
#> [35] "censReg" "cgam"
#> [37] "cgamm" "cglm"
#> [39] "clm" "clm2"
#> [41] "clmm" "clmm2"
#> [43] "clogit" "coeftest"
#> [45] "complmrob" "confusionMatrix"
#> [47] "coxme" "coxph"
#> [49] "coxph.penal" "coxph_weightit"
#> [51] "coxr" "cpglm"
#> [53] "cpglmm" "crch"
#> [55] "crq" "crqs"
#> [57] "crr" "dep.effect"
#> [59] "DirichletRegModel" "draws"
#> [61] "drc" "eglm"
#> [63] "elm" "emmGrid"
#> [65] "epi.2by2" "ergm"
#> [67] "estimate_contrasts" "estimate_means"
#> [69] "estimate_slopes" "externVar"
#> [71] "externX" "fdm"
#> [73] "feglm" "feis"
#> [75] "felm" "fitdistr"
#> [77] "fixest" "flac"
#> [79] "flexsurvreg" "flic"
#> [81] "gam" "Gam"
#> [83] "gamlss" "gamm"
#> [85] "gamm4" "garch"
#> [87] "gbm" "gee"
#> [89] "geeglm" "ggcomparisons"
#> [91] "glht" "glimML"
#> [93] "glm" "Glm"
#> [95] "glm_weightit" "glmerMod"
#> [97] "glmgee" "glmm"
#> [99] "glmmadmb" "glmmPQL"
#> [101] "glmmTMB" "glmrob"
#> [103] "glmRob" "glmx"
#> [105] "gls" "gmnl"
#> [107] "hglm" "HLfit"
#> [109] "htest" "hurdle"
#> [111] "iv_robust" "ivFixed"
#> [113] "ivprobit" "ivreg"
#> [115] "joint" "lavaan"
#> [117] "lcmm" "lm"
#> [119] "lm_robust" "lme"
#> [121] "lmerMod" "lmerModLmerTest"
#> [123] "lmodel2" "lmrob"
#> [125] "lmRob" "logistf"
#> [127] "logitmfx" "logitor"
#> [129] "logitr" "LORgee"
#> [131] "lqm" "lqmm"
#> [133] "lrm" "manova"
#> [135] "MANOVA" "marginaleffects"
#> [137] "marginaleffects.summary" "margins"
#> [139] "maxLik" "mblogit"
#> [141] "mclogit" "mcmc"
#> [143] "mcmc.list" "MCMCglmm"
#> [145] "mcp1" "mcp12"
#> [147] "mcp2" "med1way"
#> [149] "mediate" "merMod"
#> [151] "merModList" "meta_bma"
#> [153] "meta_fixed" "meta_random"
#> [155] "metaplus" "mhurdle"
#> [157] "mipo" "mira"
#> [159] "mixed" "MixMod"
#> [161] "mixor" "mjoint"
#> [163] "mle" "mle2"
#> [165] "mlm" "mlogit"
#> [167] "mmclogit" "mmlogit"
#> [169] "mmrm" "mmrm_fit"
#> [171] "mmrm_tmb" "model_fit"
#> [173] "multinom" "multinom_weightit"
#> [175] "mvord" "negbinirr"
#> [177] "negbinmfx" "nestedLogit"
#> [179] "ols" "onesampb"
#> [181] "oohbchoice" "ordinal_weightit"
#> [183] "orm" "pgmm"
#> [185] "phyloglm" "phylolm"
#> [187] "plm" "PMCMR"
#> [189] "poissonirr" "poissonmfx"
#> [191] "polr" "probitmfx"
#> [193] "psm" "Rchoice"
#> [195] "ridgelm" "riskRegression"
#> [197] "rjags" "rlm"
#> [199] "rlmerMod" "RM"
#> [201] "rma" "rma.uni"
#> [203] "rms" "robmixglm"
#> [205] "robtab" "rq"
#> [207] "rqs" "rqss"
#> [209] "rvar" "Sarlm"
#> [211] "scam" "sdmTMB"
#> [213] "selection" "sem"
#> [215] "SemiParBIV" "semLm"
#> [217] "semLme" "seqanova.svyglm"
#> [219] "serp" "slm"
#> [221] "speedglm" "speedlm"
#> [223] "stanfit" "stanmvreg"
#> [225] "stanreg" "summary.lm"
#> [227] "survfit" "survreg"
#> [229] "svy_vglm" "svy2lme"
#> [231] "svychisq" "svycoxph"
#> [233] "svyglm" "svyolr"
#> [235] "svysurvreg" "systemfit"
#> [237] "t1way" "tobit"
#> [239] "trimcibt" "truncreg"
#> [241] "vgam" "vglm"
#> [243] "wbgee" "wblm"
#> [245] "wbm" "wmcpAKP"
#> [247] "yuen" "yuend"
#> [249] "zcpglm" "zeroinfl"
#> [251] "zerotrunc"If this package helped you, please consider citing as follows:
Lüdecke D, Waggoner P, Makowski D. insight: A Unified Interface to Access Information from Model Objects in R. Journal of Open Source Software 2019;4:1412. doi: 10.21105/joss.01412
Please note that the insight project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.