
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

The Time Series Modeling Companion to healthyR
To view the full wiki, click here: Full healthyR.ts Wiki
healthyR.ts is a comprehensive R package designed
specifically for time series analysis and forecasting of hospital
administrative and clinical data. Built on the powerful tidymodels ecosystem, it provides
a consistent, user-friendly framework that simplifies complex time
series workflows.
Hospital data analysis often requires handling time series for metrics like: - Average Length of Stay (ALOS) - Readmission rates - Patient volumes and admissions - Bed occupancy rates - Clinical outcomes over time
healthyR.ts takes the guesswork out of time series
analysis by providing:
✅ Automated Workflows - One-function solutions for
complete modeling pipelines
✅ Visual Analytics - Rich plotting functions for data
exploration
✅ Data Generators - Simulate realistic time series for
testing and validation
✅ Statistical Tools - Comprehensive suite of time
series statistics
✅ Clustering - Feature-based time series clustering
capabilities
✅ Forecasting - 15 automated model workflows (ARIMA,
Prophet, XGBoost, and more)
Complete end-to-end modeling pipelines in a single function call:
Each function handles recipe creation, model specification, workflow setup, model fitting, tuning, and calibration automatically.
Generate synthetic time series data for testing: - Random walks and Brownian motion - Geometric Brownian motion - ARIMA simulations - Custom parameter configurations
Install the latest stable version from CRAN:
install.packages("healthyR.ts")Get the latest features and bug fixes from GitHub:
# install.packages("devtools")
devtools::install_github("spsanderson/healthyR.ts")Generate and visualize random walk data to understand market volatility or patient flow variations:
library(healthyR.ts)
library(ggplot2)
df <- ts_random_walk()
head(df)
#> # A tibble: 6 × 4
#> run x y cum_y
#> <dbl> <dbl> <dbl> <dbl>
#> 1 1 1 0.113 1113.
#> 2 1 2 0.119 1245.
#> 3 1 3 -0.0178 1223.
#> 4 1 4 0.141 1396.
#> 5 1 5 -0.163 1169.
#> 6 1 6 -0.0485 1112.Now that the data has been generated, lets take a look at it.
df %>%
ggplot(
mapping = aes(
x = x
, y = cum_y
, color = factor(run)
, group = factor(run)
)
) +
geom_line(alpha = 0.8) +
ts_random_walk_ggplot_layers(df)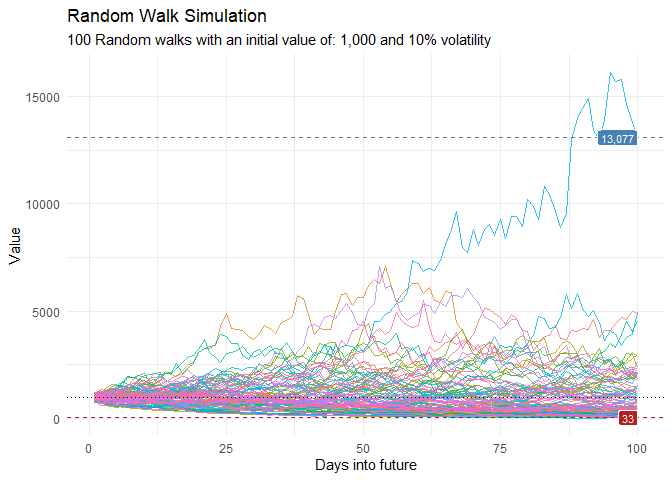
That is still pretty noisy, so lets see this in a different way. Lets clear this up a bit to make it easier to see the full range of the possible volatility of the random walks.
library(dplyr)
library(ggplot2)
df %>%
group_by(x) %>%
summarise(
min_y = min(cum_y),
max_y = max(cum_y)
) %>%
ggplot(
aes(x = x)
) +
geom_line(aes(y = max_y), color = "steelblue") +
geom_line(aes(y = min_y), color = "firebrick") +
geom_ribbon(aes(ymin = min_y, ymax = max_y), alpha = 0.2) +
ts_random_walk_ggplot_layers(df)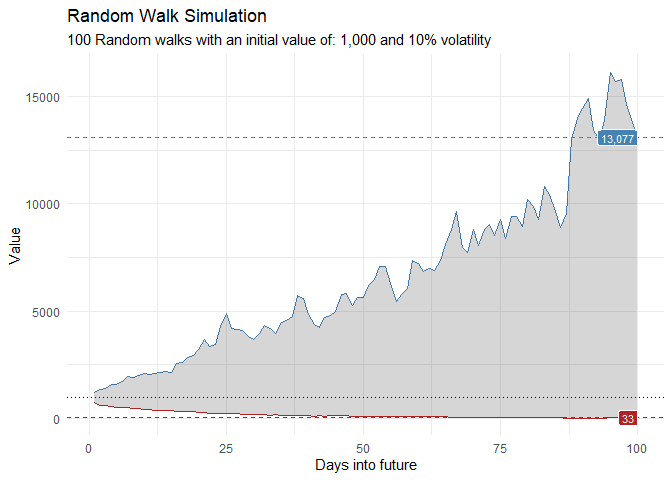
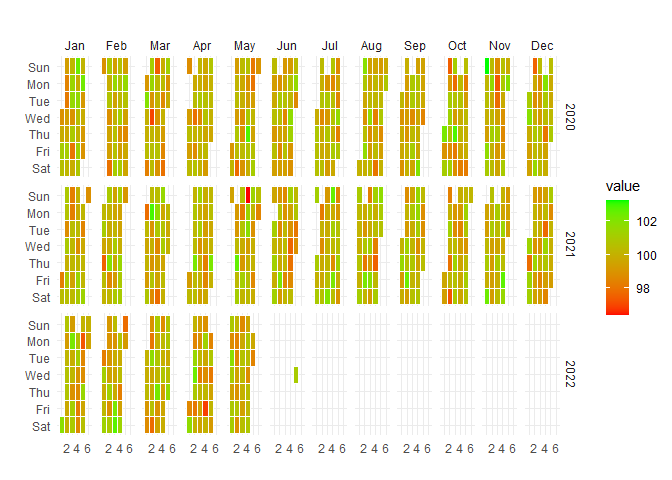
Visualize temporal patterns in your data with calendar heatmaps - perfect for identifying seasonal trends or unusual patterns in hospital metrics:
data_tbl <- data.frame(
date_col = seq.Date(
from = as.Date("2020-01-01"),
to = as.Date("2022-06-01"),
length.out = 365*2 + 180
),
value = rnorm(365*2+180, mean = 100)
)
ts_calendar_heatmap_plot(
.data = data_tbl
, .date_col = date_col
, .value_col = value
, .interactive = FALSE
)
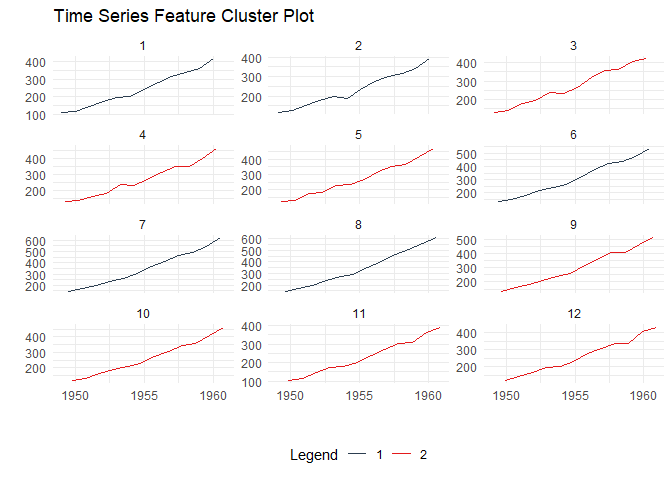
Discover patterns by clustering time series based on their statistical features:
data_tbl <- ts_to_tbl(AirPassengers) %>%
mutate(group_id = rep(1:12, 12))
output <- ts_feature_cluster(
.data = data_tbl,
.date_col = date_col,
.value_col = value,
group_id,
.features = c("acf_features","entropy"),
.scale = TRUE,
.prefix = "ts_",
.centers = 3
)
ts_feature_cluster_plot(
.data = output,
.date_col = date_col,
.value_col = value,
.center = 2,
group_id
)
#> $plot
#> $plot$static_plot
#>
#> $plot$plotly_plot
#>
#>
#> $data
#> $data$original_data
#> # A tibble: 144 × 4
#> index date_col value group_id
#> <yearmon> <date> <dbl> <int>
#> 1 Jan 1949 1949-01-01 112 1
#> 2 Feb 1949 1949-02-01 118 2
#> 3 Mar 1949 1949-03-01 132 3
#> 4 Apr 1949 1949-04-01 129 4
#> 5 May 1949 1949-05-01 121 5
#> 6 Jun 1949 1949-06-01 135 6
#> 7 Jul 1949 1949-07-01 148 7
#> 8 Aug 1949 1949-08-01 148 8
#> 9 Sep 1949 1949-09-01 136 9
#> 10 Oct 1949 1949-10-01 119 10
#> # ℹ 134 more rows
#>
#> $data$kmm_data_tbl
#> # A tibble: 3 × 3
#> centers k_means glance
#> <int> <list> <list>
#> 1 1 <kmeans> <tibble [1 × 4]>
#> 2 2 <kmeans> <tibble [1 × 4]>
#> 3 3 <kmeans> <tibble [1 × 4]>
#>
#> $data$user_item_tbl
#> # A tibble: 12 × 8
#> group_id ts_x_acf1 ts_x_acf10 ts_diff1_acf1 ts_diff1_acf10 ts_diff2_acf1
#> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.741 1.55 -0.0995 0.474 -0.182
#> 2 2 0.730 1.50 -0.0155 0.654 -0.147
#> 3 3 0.766 1.62 -0.471 0.562 -0.620
#> 4 4 0.715 1.46 -0.253 0.457 -0.555
#> 5 5 0.730 1.48 -0.372 0.417 -0.649
#> 6 6 0.751 1.61 0.122 0.646 0.0506
#> 7 7 0.745 1.58 0.260 0.236 -0.303
#> 8 8 0.761 1.60 0.319 0.419 -0.319
#> 9 9 0.747 1.59 -0.235 0.191 -0.650
#> 10 10 0.732 1.50 -0.0371 0.269 -0.510
#> 11 11 0.746 1.54 -0.310 0.357 -0.556
#> 12 12 0.735 1.51 -0.360 0.294 -0.601
#> # ℹ 2 more variables: ts_seas_acf1 <dbl>, ts_entropy <dbl>
#>
#> $data$cluster_tbl
#> # A tibble: 12 × 9
#> cluster group_id ts_x_acf1 ts_x_acf10 ts_diff1_acf1 ts_diff1_acf10
#> <int> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 2 1 0.741 1.55 -0.0995 0.474
#> 2 2 2 0.730 1.50 -0.0155 0.654
#> 3 1 3 0.766 1.62 -0.471 0.562
#> 4 1 4 0.715 1.46 -0.253 0.457
#> 5 1 5 0.730 1.48 -0.372 0.417
#> 6 2 6 0.751 1.61 0.122 0.646
#> 7 2 7 0.745 1.58 0.260 0.236
#> 8 2 8 0.761 1.60 0.319 0.419
#> 9 1 9 0.747 1.59 -0.235 0.191
#> 10 1 10 0.732 1.50 -0.0371 0.269
#> 11 1 11 0.746 1.54 -0.310 0.357
#> 12 1 12 0.735 1.51 -0.360 0.294
#> # ℹ 3 more variables: ts_diff2_acf1 <dbl>, ts_seas_acf1 <dbl>, ts_entropy <dbl>
#>
#>
#> $kmeans_object
#> $kmeans_object[[1]]
#> K-means clustering with 2 clusters of sizes 7, 5
#>
#> Cluster means:
#> ts_x_acf1 ts_x_acf10 ts_diff1_acf1 ts_diff1_acf10 ts_diff2_acf1 ts_seas_acf1
#> 1 0.7387865 1.528308 -0.2909349 0.3638392 -0.5916245 0.2930543
#> 2 0.7456468 1.568532 0.1172685 0.4858013 -0.1799728 0.2876449
#> ts_entropy
#> 1 0.6438176
#> 2 0.4918321
#>
#> Clustering vector:
#> [1] 2 2 1 1 1 2 2 2 1 1 1 1
#>
#> Within cluster sum of squares by cluster:
#> [1] 0.3660630 0.3704304
#> (between_SS / total_SS = 59.8 %)
#>
#> Available components:
#>
#> [1] "cluster" "centers" "totss" "withinss" "tot.withinss"
#> [6] "betweenss" "size" "iter" "ifault"Analyze time series behavior before and after significant events (e.g., policy changes, new treatments):
library(dplyr)
df <- ts_to_tbl(AirPassengers) %>% select(-index)
ts_time_event_analysis_tbl(
.data = df,
.horizon = 6,
.date_col = date_col,
.value_col = value,
.direction = "both"
) %>%
ts_event_analysis_plot()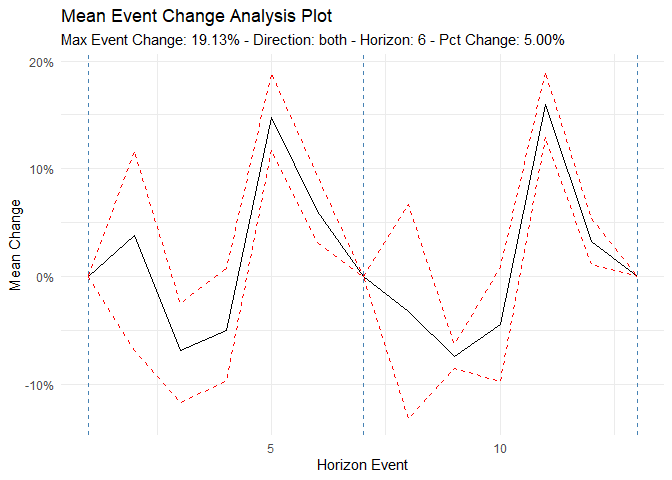
ts_time_event_analysis_tbl(
.data = df,
.horizon = 6,
.date_col = date_col,
.value_col = value,
.direction = "both"
) %>%
ts_event_analysis_plot(.plot_type = "individual")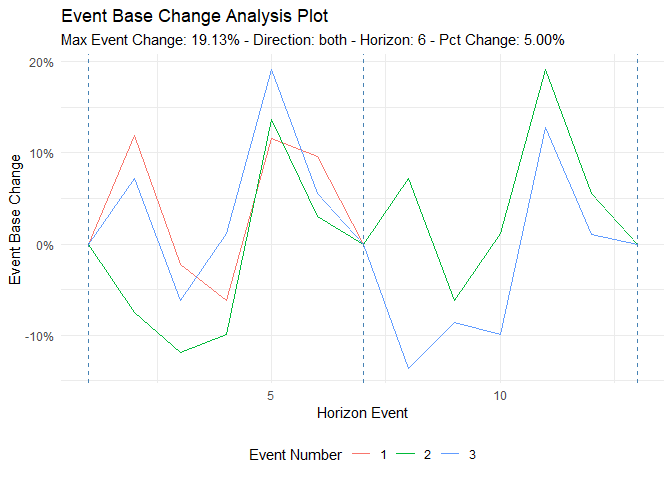
Generate realistic ARIMA time series for testing and validation:
output <- ts_arima_simulator()
output$plots$static_plot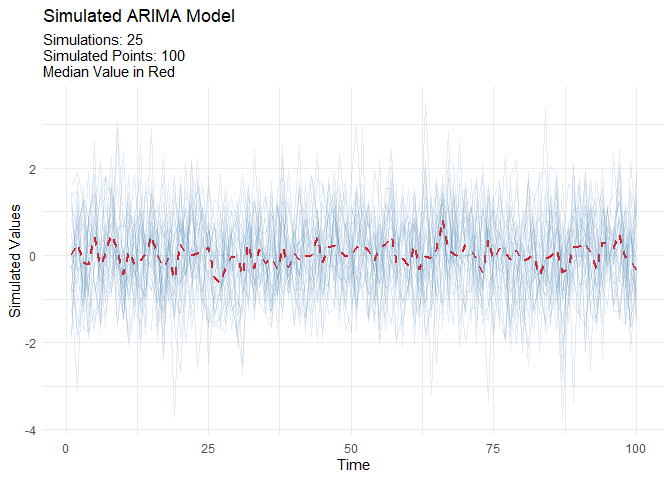
Each function creates a complete modeling pipeline including recipe, model specification, workflow, fitting, and calibration:
| Function | Model Type | Description |
|---|---|---|
ts_auto_arima() |
ARIMA | Automatic ARIMA with auto-tuning |
ts_auto_arima_xgboost() |
Hybrid | ARIMA errors with XGBoost |
ts_auto_prophet_reg() |
Prophet | Facebook’s Prophet algorithm |
ts_auto_prophet_boost() |
Hybrid | Prophet with XGBoost |
ts_auto_xgboost() |
ML | Gradient boosting |
ts_auto_nnetar() |
Neural Net | Neural network autoregression |
ts_auto_exp_smoothing() |
ETS | Exponential smoothing |
ts_auto_smooth_es() |
Smooth | Smooth package ETS |
ts_auto_theta() |
Theta | Theta method |
ts_auto_croston() |
Croston | For intermittent demand |
ts_auto_lm() |
Linear | Linear regression with time features |
ts_auto_mars() |
MARS | Multivariate adaptive regression splines |
ts_auto_glmnet() |
GLM | Elastic net regression |
ts_auto_svm_poly() |
SVM | Support vector machine (polynomial) |
ts_auto_svm_rbf() |
SVM | Support vector machine (radial) |
healthyR.ts includes 90+ functions organized into these
categories:
Contributions are welcome! Here’s how you can help:
Please follow the tidyverse style guide for code contributions.
If you use healthyR.ts in your research or publications,
please cite:
citation("healthyR.ts")Author: Steven P. Sanderson II, MPH
Maintainer: Steven P. Sanderson II, MPH
(spsanderson@gmail.com)
Copyright: © 2020-2025 Steven P. Sanderson II, MPH
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.