
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

greatR (Gene Registration from Expression and Time-courses in R) is a tool to register (align) two sets of gene expression profiles that users wish to compare.
These gene profiles data will be referred as the query and the reference data. To match the time point ranges between those profiles, the time points of the query profiles will be transformed through a stretching and shifting process. This tool uses a statistical model comparison based on a Bayesian approach to evaluate the optimality of the gene expression profiles alignment.
The flowchart below illustrates the workflow of the package given an input data:
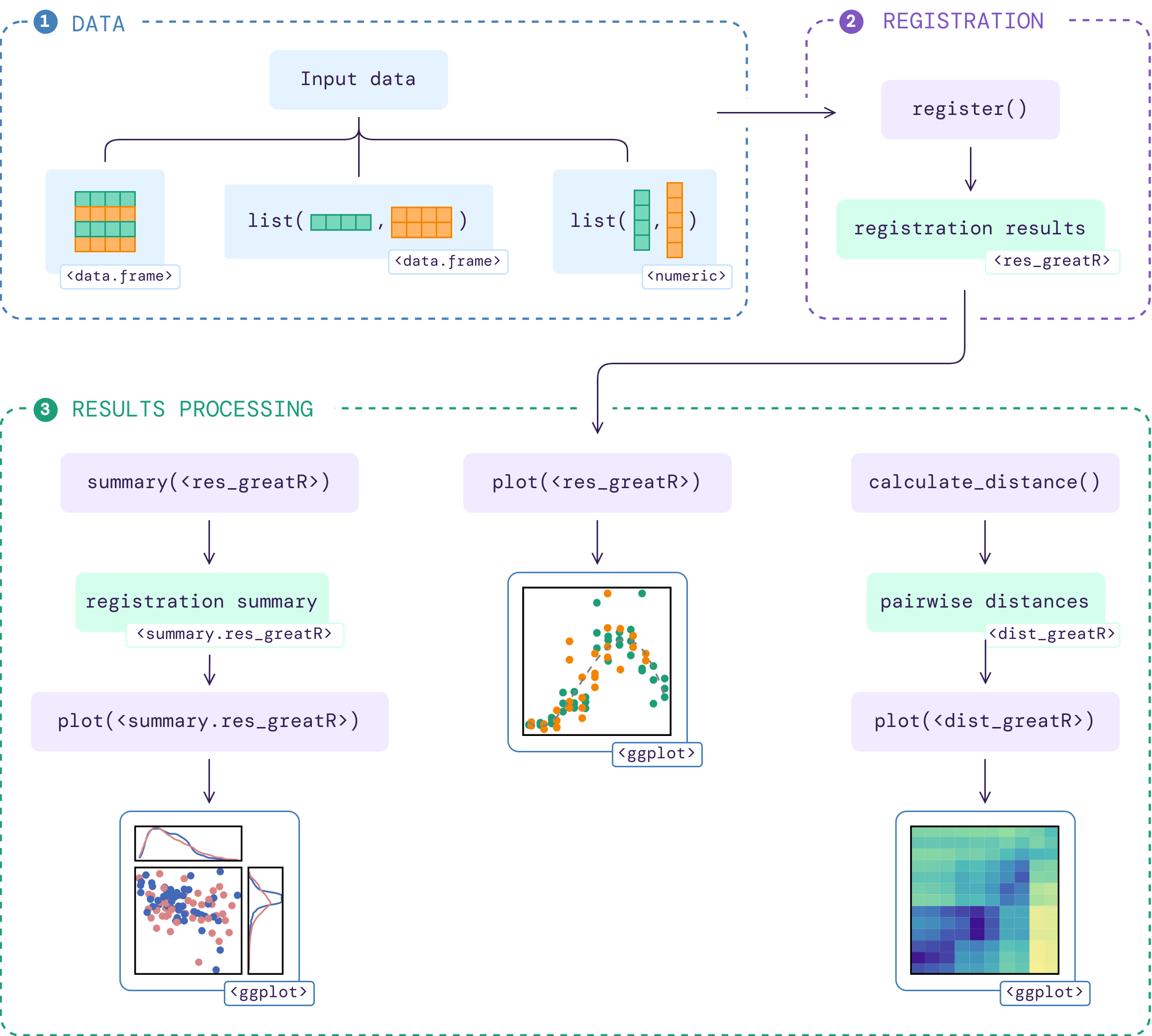
More details on how to use this package are available on function documentations and the following vignettes:
You can install the stable version of greatR from CRAN with:
And the development version of greatR from GitHub with:
This is a basic example which shows you how to register (align) gene expression profiles over time:
# Load a data frame from the sample data
b_rapa_data <- system.file("extdata/brapa_arabidopsis_data.csv", package = "greatR") |>
utils::read.csv()
# Running the registration
registration_results <- register(
b_rapa_data,
reference = "Ro18",
query = "Col0",
scaling_method = "z-score"
)
#> ── Validating input data ────────────────────────────────────────────────────────
#> ℹ Will process 10 genes.
#> ℹ Using estimated standard deviation, as no `exp_sd` was provided.
#> ℹ Using `scaling_method` = "z-score".
#>
#> ── Starting registration with optimisation ──────────────────────────────────────
#> ℹ Using L-BFGS-B optimisation method.
#> ℹ Using computed stretches and shifts search space limits.
#> ℹ Using `overlapping_percent` = 50% as a registration criterion.
#> ✔ Optimising registration parameters for genes (10/10) [2s]Calderwood, A., Hepworth, J., Woodhouse, … Morris, R. (2021). Comparative transcriptomics reveals desynchronisation of gene expression during the floral transition between Arabidopsis and Brassica rapa cultivars. Quantitative Plant Biology, 2, E4. doi:10.1017/qpb.2021.6
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.