ggmice

The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
ggmice

mice with ggplot2Enhance a mice
imputation workflow with visualizations for incomplete and/or imputed
data. The ggmice functions produce ggplot
objects which may be easily manipulated or extended. Use
ggmice to inspect missing data, develop imputation models,
evaluate algorithmic convergence, or compare observed versus imputed
data.
You can install the latest ggmice release from CRAN with:
install.packages("ggmice")Alternatively, you could install the development version of
ggmice from GitHub
with:
# install.packages("devtools")
devtools::install_github("amices/ggmice")Inspect the missing data in an incomplete dataset and subsequently
evaluate the imputed data points against observed data. See the Get started
vignette for an overview of all functionalities. Example data from mice,
showing height (in cm) by age (in years).
# load packages
library(ggplot2)
library(mice)
library(ggmice)
# load some data
dat <- boys
# visualize the incomplete data
ggmice(dat, aes(age, hgt)) + geom_point()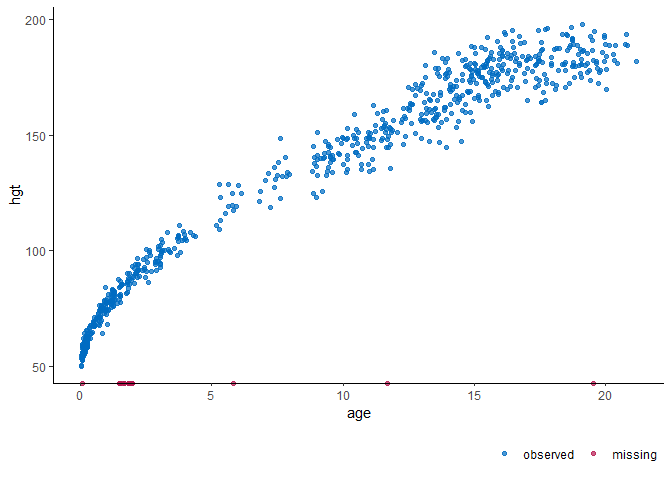
# impute the incomplete data
imp <- mice(dat, m = 1, seed = 1)
# visualize the imputed data
ggmice(imp, aes(age, hgt)) + geom_point()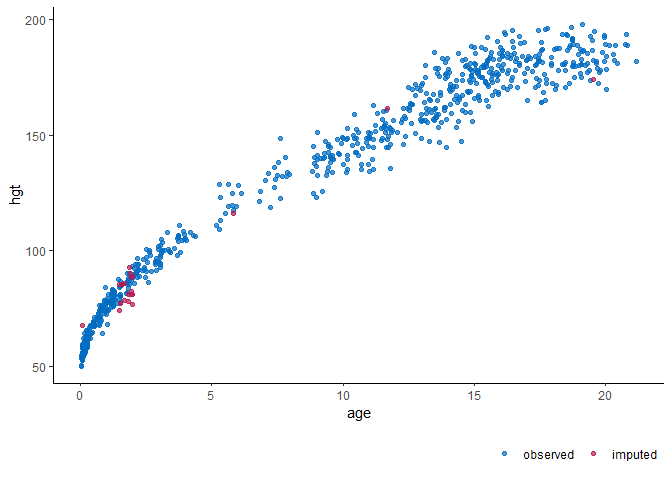
The ggmice package is developed with guidance and
feedback from the Amices team.
The ggmice hex is based on the ggplot2
and mice hex
designs.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under ReCoDID grant agreement No 825746.
You are invited to join the improvement and development of
ggmice. Please note that the project is released with a Contributor Code
of Conduct. By contributing to this project, you agree to abide by
its terms.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.