The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The ggcorrplot package can be used to visualize easily a correlation matrix using ggplot2. It provides a solution for reordering the correlation matrix and displays the significance level on the correlogram. It includes also a function for computing a matrix of correlation p-values.
Find out more at http://www.sthda.com/english/wiki/ggcorrplot-visualization-of-a-correlation-matrix-using-ggplot2.
ggcorrplot can be installed from CRAN as follow:
install.packages("ggcorrplot")Or, install the latest version from GitHub:
# Install
if(!require(devtools)) install.packages("devtools")
devtools::install_github("kassambara/ggcorrplot")# Loading
library(ggcorrplot)The mtcars data set will be used in the following R code. The function cor_pmat() [in ggcorrplot] computes a matrix of correlation p-values.
# Compute a correlation matrix
data(mtcars)
corr <- round(cor(mtcars), 1)
head(corr[, 1:6])
#> mpg cyl disp hp drat wt
#> mpg 1.0 -0.9 -0.8 -0.8 0.7 -0.9
#> cyl -0.9 1.0 0.9 0.8 -0.7 0.8
#> disp -0.8 0.9 1.0 0.8 -0.7 0.9
#> hp -0.8 0.8 0.8 1.0 -0.4 0.7
#> drat 0.7 -0.7 -0.7 -0.4 1.0 -0.7
#> wt -0.9 0.8 0.9 0.7 -0.7 1.0
# Compute a matrix of correlation p-values
p.mat <- cor_pmat(mtcars)
head(p.mat[, 1:4])
#> mpg cyl disp hp
#> mpg 0.000000e+00 6.112687e-10 9.380327e-10 1.787835e-07
#> cyl 6.112687e-10 0.000000e+00 1.802838e-12 3.477861e-09
#> disp 9.380327e-10 1.802838e-12 0.000000e+00 7.142679e-08
#> hp 1.787835e-07 3.477861e-09 7.142679e-08 0.000000e+00
#> drat 1.776240e-05 8.244636e-06 5.282022e-06 9.988772e-03
#> wt 1.293959e-10 1.217567e-07 1.222320e-11 4.145827e-05# Visualize the correlation matrix
# --------------------------------
# method = "square" (default)
ggcorrplot(corr)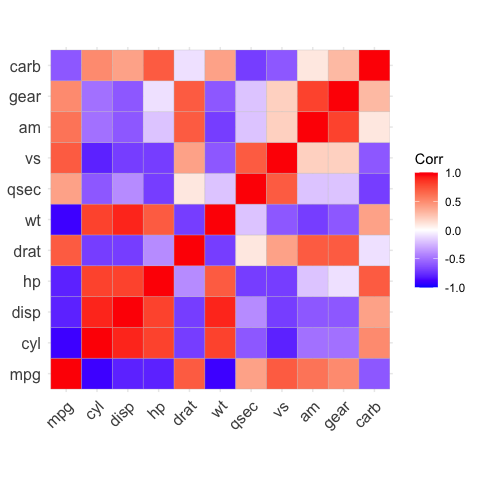
# method = "circle"
ggcorrplot(corr, method = "circle")
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> =
#> "none")` instead.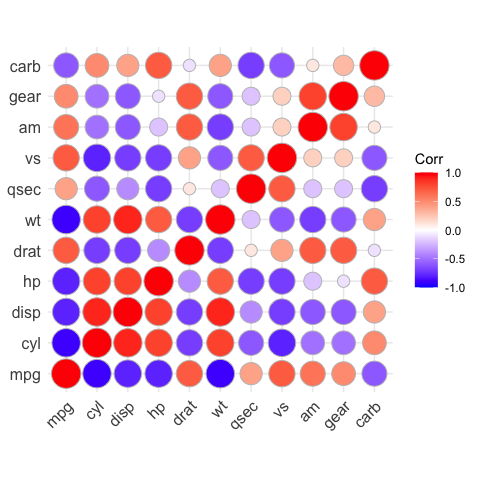
# Reordering the correlation matrix
# --------------------------------
# using hierarchical clustering
ggcorrplot(corr, hc.order = TRUE, outline.color = "white")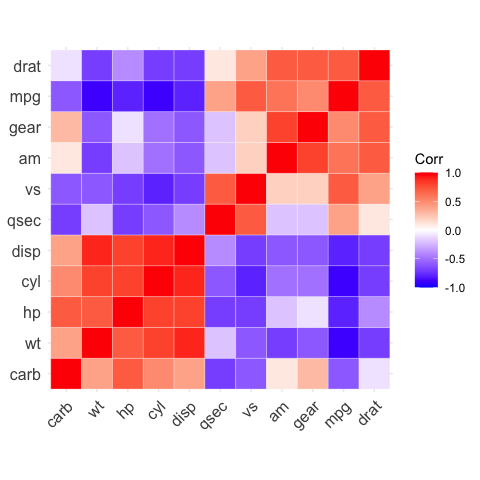
# Types of correlogram layout
# --------------------------------
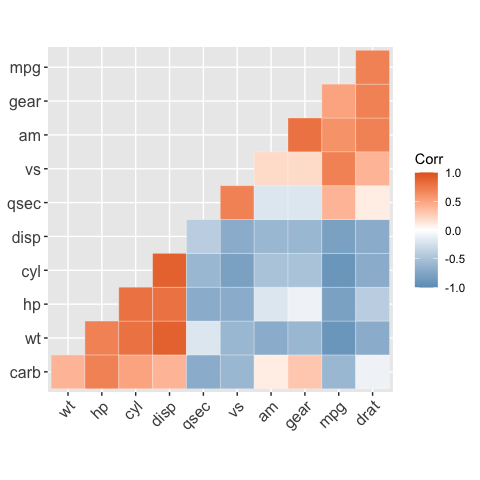
# Get the lower triangle
ggcorrplot(corr,
hc.order = TRUE,
type = "lower",
outline.color = "white")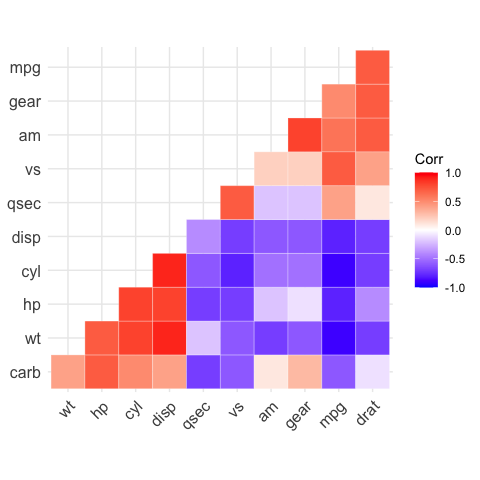
# Get the upper triangle
ggcorrplot(corr,
hc.order = TRUE,
type = "upper",
outline.color = "white")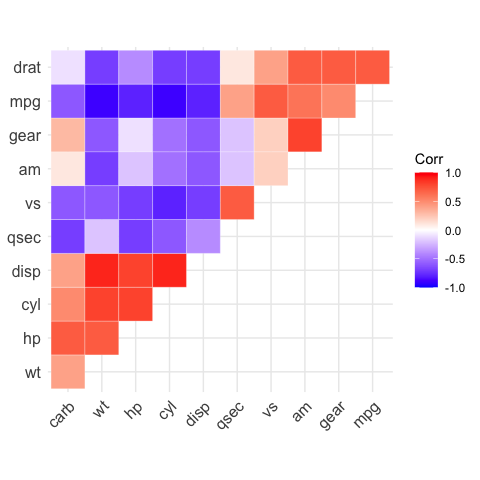
# Change colors and theme
# --------------------------------
# Argument colors
ggcorrplot(
corr,
hc.order = TRUE,
type = "lower",
outline.color = "white",
ggtheme = ggplot2::theme_gray,
colors = c("#6D9EC1", "white", "#E46726")
)
# Add correlation coefficients
# --------------------------------
# argument lab = TRUE
ggcorrplot(corr,
hc.order = TRUE,
type = "lower",
lab = TRUE)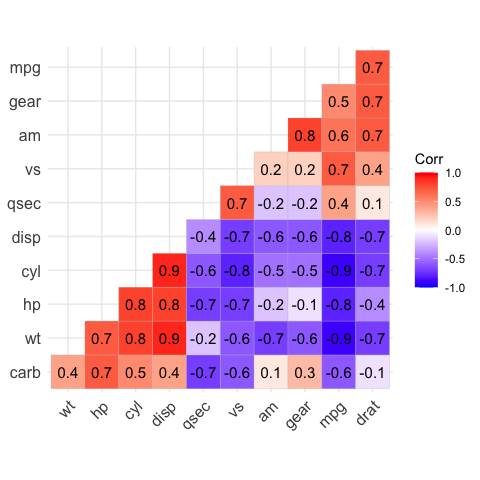
# Add correlation significance level
# --------------------------------
# Argument p.mat
# Barring the no significant coefficient
ggcorrplot(corr,
hc.order = TRUE,
type = "lower",
p.mat = p.mat)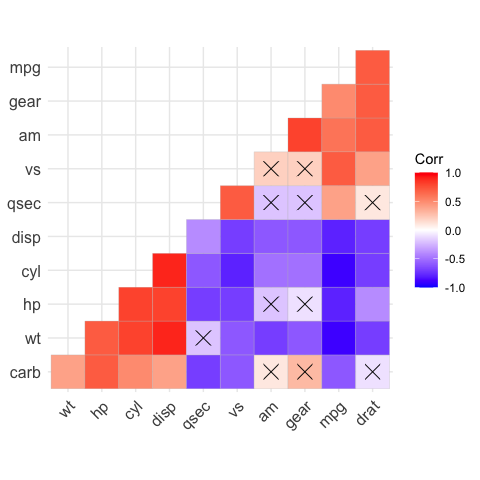
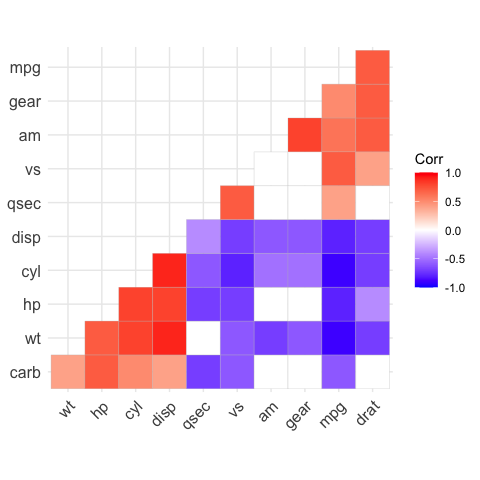
# Leave blank on no significant coefficient
ggcorrplot(
corr,
p.mat = p.mat,
hc.order = TRUE,
type = "lower",
insig = "blank"
)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.