

The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This R package implements coherent multiple imputation of general multivariate data using the GERBIL algorithm described by Robbins (2020; arXiv:2008.02243).
This package is on CRAN. You are able to install the released version of gerbil from CRAN with:
install.packages("gerbil")You can also install directly from this GitHub repository:
# run if you don't have devtools installed:
install.packages("devtools")
devtools::install_github("michaelwrobbins/gerbil")Load your dataset and run the gerbil function:
library(gerbil)
# Load the ihd sample data set with MCAR missingness:
data(ihd_mcar)
my_dataset = ihd_mcar
# Run the Imputation Process:
gerbil_object <- gerbil(dat = my_dataset, m = 1, ords = "education_level", semi = "farm_labour_days", bincat = "job_field")
#> Variable Summary:
#> Variable.Type Num.Observed Num.Miss Miss.Rate
#> sex binary 42125 0 0.00%
#> age continuous (EMP) 27382 14743 35.00%
#> marital_status binary 27382 14743 35.00%
#> job_field categorical 27382 14743 35.00%
#> farm_labour_days semicont 27382 14743 35.00%
#> own_livestock binary 27382 14743 35.00%
#> education_level ordinal 27382 14743 35.00%
#> income continuous (EMP) 27382 14743 35.00%
#>
#> Completed transformations, Time = 0.15
#> Imp. 1: gerbil initialized. Time = 2.06
#> Imp. 1: MCMC iteration 1 completed. Total time = 1.70, I-Step: 1.67, P-Step: 0.03
#> Imp. 1: MCMC iteration 2 completed. Total time = 1.81, I-Step: 1.78, P-Step: 0.03
#> Imp. 1: MCMC iteration 3 completed. Total time = 1.58, I-Step: 1.55, P-Step: 0.03
#> Imp. 1: MCMC iteration 4 completed. Total time = 1.69, I-Step: 1.67, P-Step: 0.02
#> Imp. 1: MCMC iteration 5 completed. Total time = 1.69, I-Step: 1.66, P-Step: 0.03
#> Imp. 1: MCMC iteration 6 completed. Total time = 1.75, I-Step: 1.72, P-Step: 0.03
#> Imp. 1: MCMC iteration 7 completed. Total time = 1.70, I-Step: 1.67, P-Step: 0.03
#> Imp. 1: MCMC iteration 8 completed. Total time = 1.66, I-Step: 1.63, P-Step: 0.03
#> Imp. 1: MCMC iteration 9 completed. Total time = 1.71, I-Step: 1.68, P-Step: 0.03
#> Imp. 1: MCMC iteration 10 completed. Total time = 1.53, I-Step: 1.50, P-Step: 0.03
#> Imp. 1: MCMC iteration 11 completed. Total time = 1.77, I-Step: 1.73, P-Step: 0.04
#> Imp. 1: MCMC iteration 12 completed. Total time = 1.66, I-Step: 1.63, P-Step: 0.03
#> Imp. 1: MCMC iteration 13 completed. Total time = 1.74, I-Step: 1.71, P-Step: 0.03
#> Imp. 1: MCMC iteration 14 completed. Total time = 1.78, I-Step: 1.75, P-Step: 0.03
#> Imp. 1: MCMC iteration 15 completed. Total time = 1.78, I-Step: 1.75, P-Step: 0.03
#> Imp. 1: MCMC iteration 16 completed. Total time = 1.67, I-Step: 1.64, P-Step: 0.03
#> Imp. 1: MCMC iteration 17 completed. Total time = 1.86, I-Step: 1.83, P-Step: 0.03
#> Imp. 1: MCMC iteration 18 completed. Total time = 1.61, I-Step: 1.58, P-Step: 0.03
#> Imp. 1: MCMC iteration 19 completed. Total time = 1.55, I-Step: 1.51, P-Step: 0.04
#> Imp. 1: MCMC iteration 20 completed. Total time = 1.64, I-Step: 1.61, P-Step: 0.03
#> Imp. 1: MCMC iteration 21 completed. Total time = 1.57, I-Step: 1.54, P-Step: 0.03
#> Imp. 1: MCMC iteration 22 completed. Total time = 1.58, I-Step: 1.55, P-Step: 0.03
#> Imp. 1: MCMC iteration 23 completed. Total time = 1.52, I-Step: 1.49, P-Step: 0.03
#> Imp. 1: MCMC iteration 24 completed. Total time = 1.55, I-Step: 1.53, P-Step: 0.02
#> Imp. 1: MCMC iteration 25 completed. Total time = 1.49, I-Step: 1.47, P-Step: 0.02
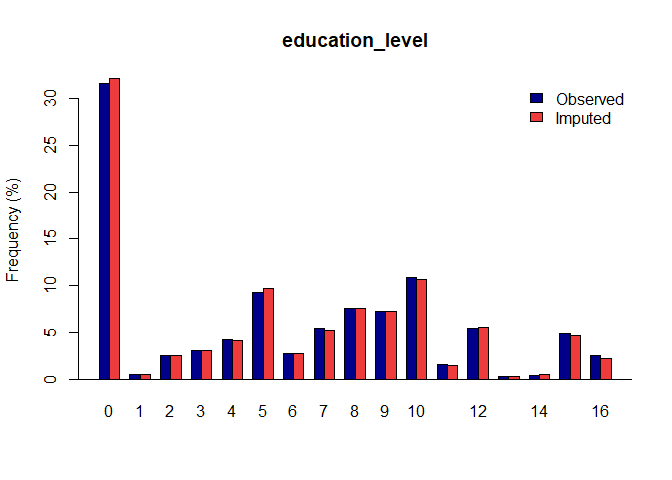
#> Completed untransformations for imputed dataset 1, Time = 0.04Once you have a gerbil object, you can use the plot
function to verify the quality of your imputations:
plot(gerbil_object)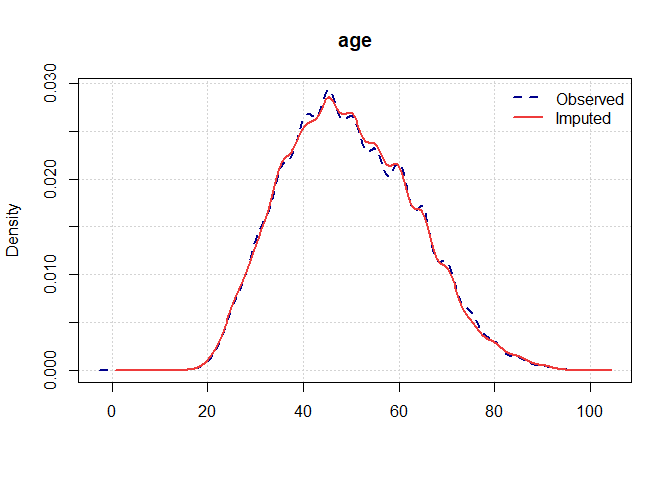





We have developed package vignettes that are available within the ./vignettes folder in this repository.
This package is tested at every build by the automated tests listed
within the ./tests/testthat folder.
One can verify our test coverage statistics by opening the r package r project and running:
# load all functions
devtools::test_coverage()These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.