The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This package implements the GenSVM Multiclass Support Vector Machine classifier in R.
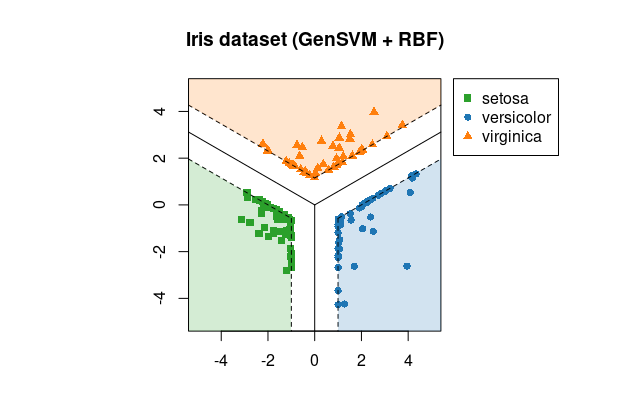
# Plot created with:
> library(gensvm)
> x <- iris[, -5]
> y <- iris[, 5]
> fit <- gensvm(x, y, kernel='rbf', gamma=10, max.iter=5000, verbose=1, random.seed=123)
> plot(fit, xlim=c(-5, 5), ylim=c(-5, 5))
> title("Iris dataset (GenSVM + RBF)")The GenSVM classifier is a generalized multiclass support vector machine (SVM). This classifier aims to find decision boundaries that separate the classes with as wide a margin as possible. In GenSVM, the loss functions that measures how misclassifications are counted is very flexible. This allows the user to tune the classifier to the dataset at hand and potentially obtain higher classification accuracy. Moreover, this flexibility means that GenSVM has a number of alternative multiclass SVMs as special cases. One of the other advantages of GenSVM is that it is trained in the primal space, allowing the use of warm starts during optimization. This means that for common tasks such as cross validation or repeated model fitting, GenSVM can be trained very quickly.
For more information about GenSVM, see the paper: GenSVM: A Generalized Multiclass Support Vector Machine by G.J.J. van den Burg and P.J.F. Groenen (Journal of Machine Learning Research, 2016).
This package can be installed from CRAN:
> install.packages('gensvm')The package is extensively documented with many examples. See
?gensvm-package, ?gensvm and
?gensvm.grid in R.
The main GenSVM functions are: * gensvm : Fit a GenSVM
model for specific model parameters. * gensvm.grid : Run a
cross-validated grid search for GenSVM.
Both these functions return S3 objects for which plot
and predict functions are available. For the GenSVMGrid
object the function is applied to the best model found during training.
For both of these objects a coef function is also
available.
The following utility functions are also included in the package: *
gensvm.accuracy : Compute the accuracy score between true
and predicted class labels * gensvm.maxabs.scale : Scale
each column of the dataset by its maximum absolute value, preserving
sparsity and mapping the data to [-1, 1] *
gensvm.train.test.split : Split a dataset into a training
and testing sample * gensvm.refit : Refit a fitted GenSVM
model with slightly different parameters or on a different dataset
If you use GenSVM in your work, please cite the paper using the information available through the following R command:
> citation('gensvm')Alternatively, you can use the following BibTeX code directly:
@article{JMLR:v17:14-526,
author = {Gerrit J.J. {van den Burg} and Patrick J.F. Groenen},
title = {{GenSVM}: A Generalized Multiclass Support Vector Machine},
journal = {Journal of Machine Learning Research},
year = {2016},
volume = {17},
number = {225},
pages = {1-42},
url = {https://jmlr.org/papers/v17/14-526.html}
}Copyright 2018, G.J.J. van den Burg.
RGenSVM is free software: you can redistribute it and/or modify
it under the terms of the GNU General Public License as published by
the Free Software Foundation, either version 3 of the License, or
(at your option) any later version.
RGenSVM is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU General Public License for more details.
You should have received a copy of the GNU General Public License
along with RGenSVM. If not, see <http://www.gnu.org/licenses/>.
For more information please contact:
G.J.J. van den Burg
email: gertjanvandenburg@gmail.comThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.