
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Determining the convergence of Markov Chain Monte Carlo (MCMC) algorithms run on highly-dimensional or un-ordered spaces is an active area of research. This package implements several distance based algorithms for the creation of diagnostics in these situations. A forthcoming paper describes the methodology used in detail.
#Install from CRAN
install.packages('genMCMCDiag')
#Install from gitHub
# install.packages("devtools")
devtools::install_github("LukeDuttweiler/genMCMCDiag")Consider the results of a multi-chain MCMC algorithm over a
univariate space. The genMCMCDiag package provides a simple
interface to retrieve traceplots, the effective sample size (ESS), and
the Gelman-Rubin Potential Scale Reduction Factor (PSRF).
We demonstrate the usage of genMCMCDiag in this simple
scenario, using the included simulated MCMC results
uniMCMCResults. The output of genDiagnostic is
a traceplot and tables containing the ESS and PSRF results. The method
is set to ‘standard’ here as we don’t want to transform the data prior
to running the diagnostics, as it is already univariate.
#Load package
library(genMCMCDiag)
#View structure of uniMCMCResults
str(uniMCMCResults)
#> List of 7
#> $ : num [1:2000] 5.72 5.72 5.72 -5.67 -5.67 ...
#> $ : num [1:2000] -3.98 -3.98 -3.98 3.81 3.61 ...
#> $ : num [1:2000] 2.17 2.34 -2.35 -2.35 -2.42 ...
#> $ : num [1:2000] 0.1266 0.058 0.0436 0.0103 0.0194 ...
#> $ : num [1:2000] 2 2 2 2 2 ...
#> $ : num [1:2000] 3.86 -3.74 3.75 3.75 3.75 ...
#> $ : num [1:2000] 5.87 -5.83 5.81 5.81 5.81 ...
#View Diagnostics
genDiagnostic(uniMCMCResults, proximityMap = 'standard')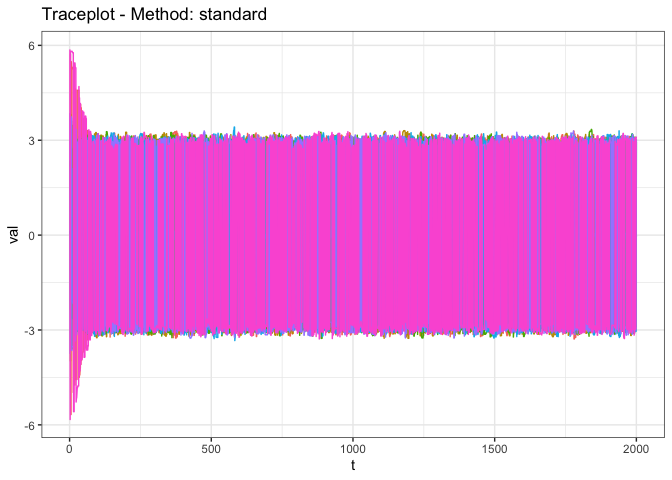
#> ----------------------------------------------------
#> Generalized MCMC Diagnostics using Custom Method
#> ----------------------------------------------------
#>
#> |Effective Sample Size:
#> |---------------------------
#> | Chain 1| Chain 2| Chain 3| Chain 4| Chain 5| Chain 6| Chain 7| Sum|
#> |-------:|-------:|-------:|-------:|-------:|--------:|--------:|--------:|
#> | 847.966| 908.501| 885.617| 828.86| 694.21| 1052.483| 1019.646| 6237.283|
#>
#> |psrf:
#> |---------------------------
#> | Point est.| Upper C.I.|
#> |----------:|----------:|
#> | 1.014| 1.015|Consider the results of a more complex multi-chain MCMC algorithm run on the space of Bayesian network (BN) partitions. The space of BN partitions does not have a natural distance based order (unlike the real-line used in the previous example), and so standard traceplots and MCMC diagnostics cannot be used.
genMCMCDiag includes transformation methods to help deal
with this issue, based on a user-supplied (or built in) distance
function. In this instance we will use the built-in
partitionDistance and the ts transformation
method (details to be published in a forthcoming paper), to view a
diagnostic traceplot and transformed calculations of the ESS and PSRF
diagnostics.
#View Diagnostics from ts method
genDiagnostic(bnMCMCResults, proximityMap = 'ts', distance = partitionDist)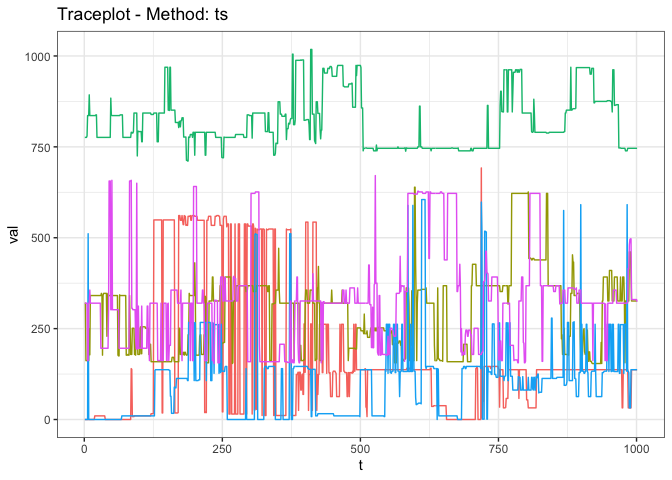
#> ----------------------------------------------------
#> Generalized MCMC Diagnostics using Custom Method
#> ----------------------------------------------------
#>
#> |Effective Sample Size:
#> |---------------------------
#> | Chain 1| Chain 2| Chain 3| Chain 4| Chain 5| Sum|
#> |-------:|-------:|-------:|-------:|-------:|------:|
#> | 30.553| 25.257| 18.956| 74.901| 32.204| 181.87|
#>
#> |psrf:
#> |---------------------------
#> | Point est.| Upper C.I.|
#> |----------:|----------:|
#> | 2.921| 5.161|The diagnostic results clearly reveal a complete lack of mixing!
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.