The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

The goal of functiondepends is to allow for tidy exploration of unstructured codebase without evaluation of code.
One can install functiondepends from CRAN:
install.packages("functiondepends")or development version from GitHub:
# install.packages("devtools")
devtools::install_github("jakubsob/functiondepends")library(functiondepends)
# Create environment for loaded functions
envir <- new.env()
# Search recursively current directory
functions <- find_functions(".", envir = envir, recursive = TRUE)functions
#> # A tibble: 5 × 3
#> Path Function SourceFile
#> <chr> <chr> <chr>
#> 1 R find_dependencies find-dependencies.R
#> 2 R is_function find-functions.R
#> 3 R get_function_name find-functions.R
#> 4 R is_assign find-functions.R
#> 5 R find_functions find-functions.RSearch for dependencies of function find_functions
within parsed functions:
dependency <- find_dependencies("find_functions", envir = envir, in_envir = TRUE)
dependency
#> # A tibble: 2 × 5
#> Source SourceRep SourceNamespace Target TargetInDegree
#> <chr> <int> <chr> <chr> <int>
#> 1 get_function_name 1 user-defined find_functions 2
#> 2 is_function 1 user-defined find_functions 2Note that SourceNamespace column has value
user-defined as the functions are searched within source of
the package.
Search for all dependencies of find_functions
function:
library(ggplot2)
library(dplyr)
dependency <- find_dependencies("find_functions", envir = envir, in_envir = FALSE)
dependency %>%
slice_max(SourceRep, n = 10) %>%
mutate(Source = reorder(Source, SourceRep)) %>%
ggplot(aes(x = Source, y = SourceRep, fill = SourceNamespace)) +
geom_col() +
coord_flip() +
labs(caption = "Top 10 most repeated calls in 'find_functions'.")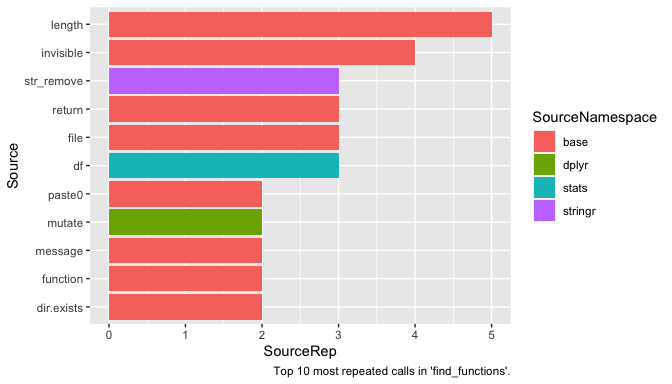
Note that name df is often used to store object of type
data.frame. df is also a name of F
distribution density function from stats package. If you
suspect that given function ought not to use a specific package, see the
source code of function to check the context. To do so, one can execute
find_dependencies function with add_info
argument set to TRUE.
library(tidyr)
dependency <- find_dependencies("find_functions", envir = envir, in_envir = FALSE, add_info = TRUE)
dependency %>%
filter(SourceNamespace == "stats") %>%
select(Source, SourcePosition, SourceContext) %>%
unnest(c(SourcePosition, SourceContext))
#> # A tibble: 6 × 3
#> Source SourcePosition SourceContext
#> <chr> <dbl> <chr>
#> 1 df 10 " df <- purrr::map_dfr(sourceFiles, function(file) {"
#> 2 df 19 " source_name <- basename(df$Path)"
#> 3 df 21 " df <- df %>% dplyr::mutate(Path = stringr::str_rem…
#> 4 df 23 " paths <- stringr::str_split(df$Path, \"/|\\\\\…
#> 5 df 25 " df <- tidyr::separate(df, \"Path\", into = pas…
#> 6 df 27 " df %>% dplyr::mutate(SourceFile = source_name)"One can see that indeed df is not a call to function
stats::df.
dependency <- find_dependencies(unique(functions$Function), envir = envir, in_envir = FALSE)
dependency %>%
distinct(Target, TargetInDegree) %>%
mutate(Target = reorder(Target, TargetInDegree)) %>%
ggplot(aes(x = Target, y = TargetInDegree)) +
geom_col() +
coord_flip() +
labs(caption = "Functions with most function calls.")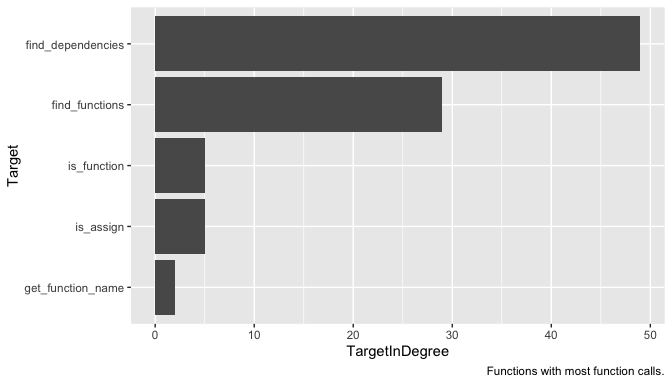
dependency <- find_dependencies(unique(functions$Function), envir = envir, in_envir = FALSE)
dependency %>%
group_by(SourceNamespace) %>%
tally(name = "Count") %>%
slice_max(Count, n = 10) %>%
mutate(SourceNamespace = reorder(SourceNamespace, Count)) %>%
ggplot(aes(x = SourceNamespace, y = Count)) +
geom_col() +
coord_flip() +
labs(caption = "Top 10 used namespaces.")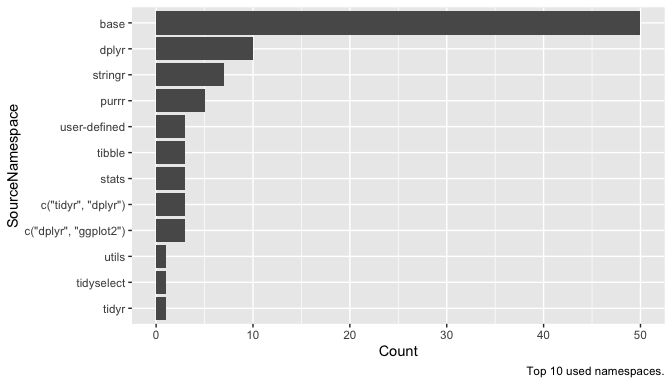
See which user-defined functions depend most on other user-defined functions within searched codebase.
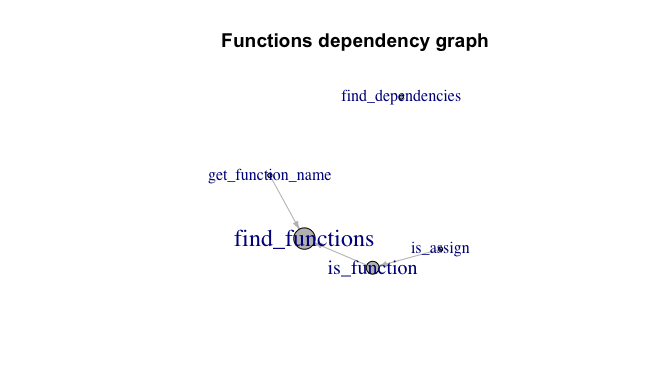
dependency <- find_dependencies(unique(functions$Function), envir = envir, in_envir = TRUE)
dependency %>%
distinct(Target, TargetInDegree) %>%
arrange(-TargetInDegree)
#> # A tibble: 5 × 2
#> Target TargetInDegree
#> <chr> <dbl>
#> 1 find_functions 2
#> 2 is_function 1
#> 3 find_dependencies 0
#> 4 get_function_name 0
#> 5 is_assign 0library(igraph)
edges <- dependency %>%
select(Source, Target) %>%
na.omit()
vertices <- unique(c(dependency$Source, dependency$Target))
vertices <- vertices[!is.na(vertices)]
g <- graph_from_data_frame(d = edges, vertices = vertices)
deg <- degree(g, mode = "in")
V(g)$size <- deg * 10 + 5
V(g)$label.cex <- (degree(g, mode = "in", normalized = TRUE) + 1)
plot(
g,
vertex.color = "grey",
edge.color = "grey",
edge.arrow.size = .4,
main = "Functions dependency graph"
)
dependency <- find_dependencies(unique(functions$Function), envir = envir, in_envir = FALSE)
edges <- dependency %>%
select(Source, Target) %>%
na.omit()
vertices <- unique(c(edges$Source, edges$Target))
g <- graph_from_data_frame(edges)
deg <- degree(g, mode = "in")
V(g)$size <- deg
V(g)$label.cex <- (degree(g, mode = "in", normalized = TRUE) + 1) / 1.8
plot(
g,
vertex.color = "grey",
edge.color = "grey",
edge.arrow.size = .4,
main = "Full functions dependency graph"
)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.