
Example data with heterogeneous treatment effects
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The goal of did2s is to estimate TWFE models without running into the problem of staggered treatment adoption.
For common issues, see this issue: https://github.com/kylebutts/did2s/issues/12
You can install did2s from CRAN with:
install.packages("did2s")To install the development version, run the following:
devtools::install_github("kylebutts/did2s")For details on the methodology, view this vignette
To view the documentation, type ?did2s into the
console.
The main function is did2s which estimates the two-stage
did procedure. This function requires the following options:
yname: the outcome variablefirst_stage: formula for first stage, can include fixed
effects and covariates, but do not include treatment variable(s)!second_stage: This should be the treatment variable or
in the case of event studies, treatment variables.treatment: This has to be the 0/1 treatment variable
that marks when treatment turns on for a unit. If you suspect
anticipation, see note above for accounting for this.cluster_var: Which variables to cluster onOptional options:
weights: Optional variable to run a weighted first- and
second-stage regressionsbootstrap: Should standard errors be bootstrapped
instead? Default is False.n_bootstraps: How many clustered bootstraps to perform
for standard errors. Default is 250.did2s returns a list with two objects:
I will load example data from the package and plot the average outcome among the groups.
# Automatically loads fixest
library(did2s)
#> Loading required package: fixest
#> did2s (v1.1.0). For more information on the methodology, visit <https://www.kylebutts.github.io/did2s>
#>
#> To cite did2s in publications use:
#>
#> Butts & Gardner, "The R Journal: did2s: Two-Stage
#> Difference-in-Differences", The R Journal, 2022
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {did2s: Two-Stage Difference-in-Differences Following Gardner (2021)},
#> author = {Kyle Butts and John Gardner},
#> year = {2021},
#> url = {https://journal.r-project.org/articles/RJ-2022-048/},
#> }
# Load Data from R package
data("df_het", package = "did2s")
df_het = as.data.frame(df_het)Here is a plot of the average outcome variable for each of the groups:
# Mean for treatment group-year
agg <- aggregate(df_het$dep_var, by = list(g = df_het$g, year = df_het$year), FUN = mean)
agg$g <- as.character(agg$g)
agg$g <- ifelse(agg$g == "0", "Never Treated", agg$g)
never <- agg[agg$g == "Never Treated", ]
g1 <- agg[agg$g == "2000", ]
g2 <- agg[agg$g == "2010", ]
plot(0, 0,
xlim = c(1990, 2020), ylim = c(3.5, 7.2), type = "n",
main = "Data-generating Process", ylab = "Outcome", xlab = "Year"
)
abline(v = c(1999.5, 2009.5), lty = 2)
lines(never$year, never$x, col = "#8e549f", type = "b", pch = 15)
lines(g1$year, g1$x, col = "#497eb3", type = "b", pch = 17)
lines(g2$year, g2$x, col = "#d2382c", type = "b", pch = 16)
legend(
x = 1990, y = 7.1, col = c("#8e549f", "#497eb3", "#d2382c"),
pch = c(15, 17, 16),
legend = c("Never Treated", "2000", "2010")
)
Example data with heterogeneous treatment effects
First, lets estimate a static did. There are two things to note here.
First, note that I can use fixest::feols formula including
the | for specifying fixed effects and
fixest::i for improved factor variable support. Second,
note that did2s returns a fixest estimate
object, so fixest::etable, fixest::coefplot,
and fixest::iplot all work as expected.
# Static
static <- did2s(
df_het,
yname = "dep_var", first_stage = ~ 0 | state + year,
second_stage = ~ i(treat, ref = FALSE), treatment = "treat",
cluster_var = "state"
)
#> Running Two-stage Difference-in-Differences
#> - first stage formula `~ 0 | state + year`
#> - second stage formula `~ i(treat, ref = FALSE)`
#> - The indicator variable that denotes when treatment is on is `treat`
#> - Standard errors will be clustered by `state`
fixest::etable(static)
#> static
#> Dependent Var.: dep_var
#>
#> treat = TRUE 2.152*** (0.0476)
#> _______________ _________________
#> S.E. type Custom
#> Observations 46,500
#> R2 0.33790
#> Adj. R2 0.33790
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1This is very close to the true treatment effect of ~2.23.
Then, let’s estimate an event study did. Note that relative year has
a value of Inf for never treated, so I put this as a
reference in the second stage formula.
# Event Study
es <- did2s(df_het,
yname = "dep_var", first_stage = ~ 0 | state + year,
second_stage = ~ i(rel_year, ref = Inf), treatment = "treat",
cluster_var = "state"
)
#> Running Two-stage Difference-in-Differences
#> - first stage formula `~ 0 | state + year`
#> - second stage formula `~ i(rel_year, ref = Inf)`
#> - The indicator variable that denotes when treatment is on is `treat`
#> - Standard errors will be clustered by `state`And plot the results:
fixest::iplot(es, main = "Event study: Staggered treatment", xlab = "Relative time to treatment", col = "steelblue", ref.line = -0.5, drop = "Inf")
# Add the (mean) true effects
true_effects <- head(tapply((df_het$te + df_het$te_dynamic), df_het$rel_year, mean), -1)
points(-20:20, true_effects, pch = 20, col = "black")
# Legend
legend(
x = -20, y = 3, col = c("steelblue", "black"), pch = c(20, 20),
legend = c("Two-stage estimate", "True effect")
)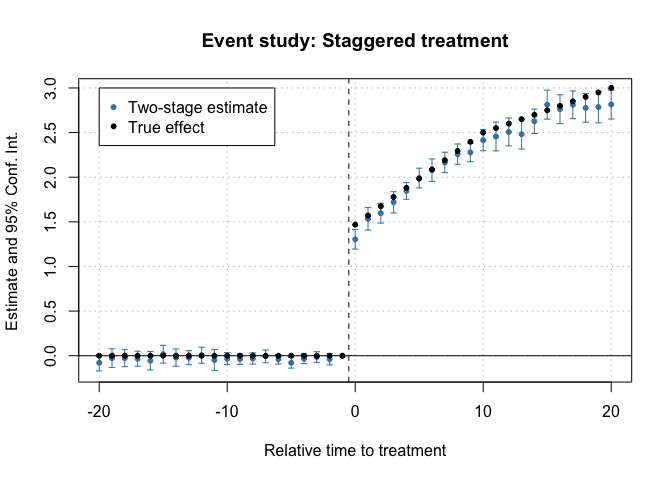
Event-study plot with example data
twfe <- feols(dep_var ~ i(rel_year, ref = c(Inf, -1)) | unit + year, data = df_het)
fixest::iplot(
list(es, twfe),
sep = 0.2, ref.line = -0.5,
col = c("steelblue", "#82b446"), pt.pch = c(20, 18),
xlab = "Relative time to treatment",
main = "Event study: Staggered treatment (comparison)",
drop = "Inf"
)
# Legend
legend(
x = -20, y = 3, col = c("steelblue", "#82b446"), pch = c(20, 18),
legend = c("Two-stage estimate", "TWFE")
)
TWFE and Two-Stage estimates of Event-Study
In version 1.1.0, we added support for computing a sensitivity analysis using the approach of Rambachan and Roth (2021).
Here’s an example using data from here.
The provided dataset ehec_data.dta contains a state-level
panel dataset on health insurance coverage and Medicaid expansion. The
variable dins shows the share of low-income childless
adults with health insurance in the state. The variable
yexp2 gives the year that a state expanded Medicaid
coverage under the Affordable Care Act, and is missing if the state
never expanded.
library(HonestDiD)
library(ggplot2)
df <- haven::read_dta("https://raw.githubusercontent.com/Mixtape-Sessions/Advanced-DID/main/Exercises/Data/ehec_data.dta")
df$treated <- ifelse(is.na(df$yexp2), 0, 1 * (df$year >= df$yexp2))
df$rel_year <- ifelse(is.na(df$yexp2), -100, df$year - df$yexp2)
# Estimate did2s
es_did2s <- did2s(
df,
yname = "dins",
first_stage = ~ 0 | stfips + year,
second_stage = ~ 0 + i(rel_year, ref = -100),
treatment = "treated",
cluster_var = "stfips"
)
#> Running Two-stage Difference-in-Differences
#> - first stage formula `~ 0 | stfips + year`
#> - second stage formula `~ 0 + i(rel_year, ref = -100)`
#> - The indicator variable that denotes when treatment is on is `treated`
#> - Standard errors will be clustered by `stfips`
iplot(es_did2s, drop = "-100")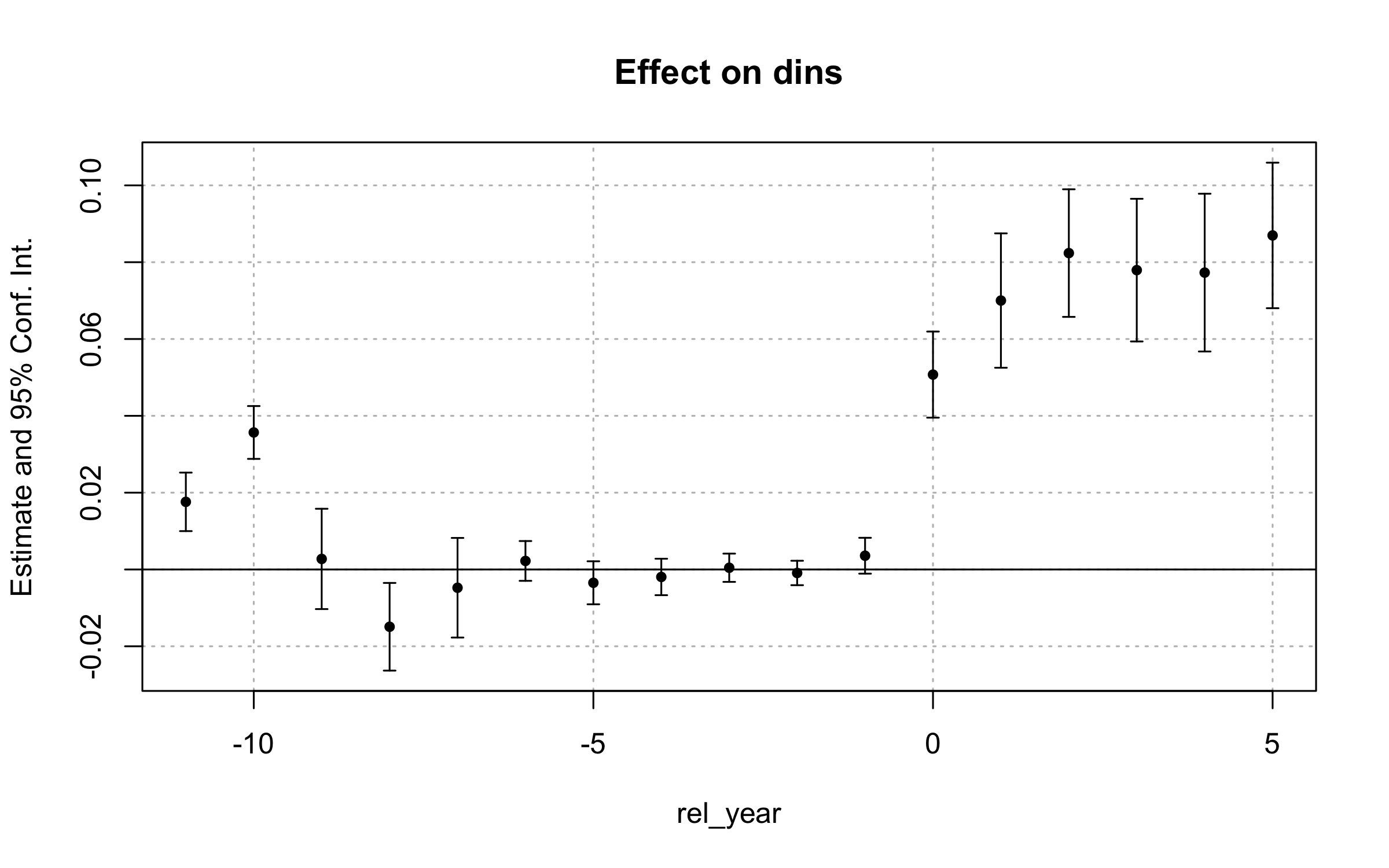
Estimates of the effect of Medicaid expansion on health insurance coverage
# Relative Magnitude sensitivity analysis
sensitivity_results <- es_did2s |>
# Take fixest obj and convert for `honest_did_did2s`
get_honestdid_obj_did2s(coef_name = "rel_year") |>
# Run sensitivity analysis
honest_did_did2s(
e = 0,
type = "relative_magnitude",
Mbarvec = seq(from = 0.5, to = 2, by = 0.5)
)
#> Warning in .ARP_computeCI(betahat = betahat, sigma = sigma, numPrePeriods =
#> numPrePeriods, : CI is open at one of the endpoints; CI length may not be
#> accurate
#> Warning in .ARP_computeCI(betahat = betahat, sigma = sigma, numPrePeriods =
#> numPrePeriods, : CI is open at one of the endpoints; CI length may not be
#> accurate
#> Warning in .ARP_computeCI(betahat = betahat, sigma = sigma, numPrePeriods =
#> numPrePeriods, : CI is open at one of the endpoints; CI length may not be
#> accurate
# Create plot
HonestDiD::createSensitivityPlot_relativeMagnitudes(
sensitivity_results$robust_ci,
sensitivity_results$orig_ci
)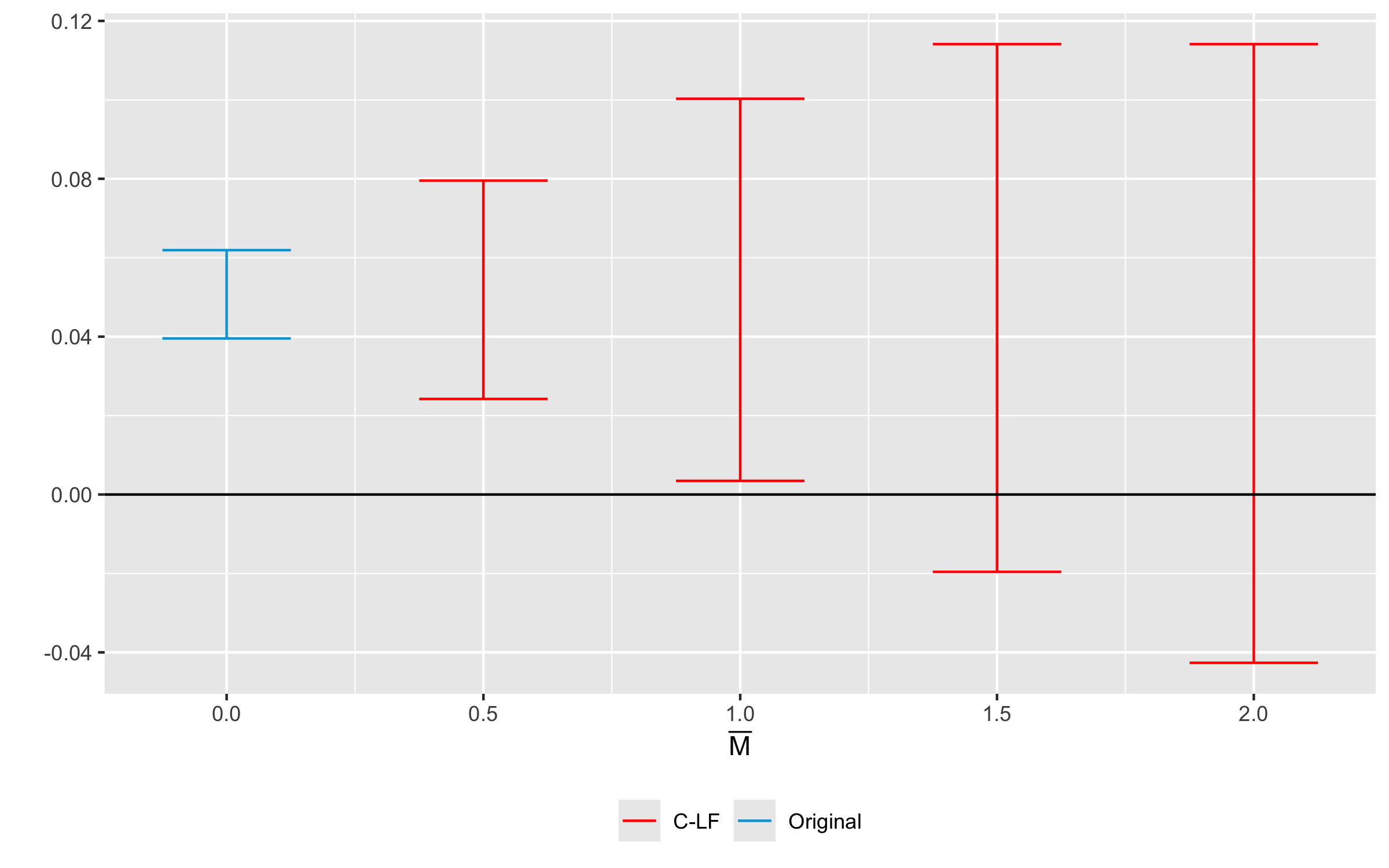
Sensitivity analysis for the example data
library(tidyverse)
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.5
#> ✔ forcats 1.0.0 ✔ stringr 1.5.1
#> ✔ lubridate 1.9.3 ✔ tibble 3.2.1
#> ✔ purrr 1.0.2 ✔ tidyr 1.3.1
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsdata(df_het)
df = df_het
multiple_ests = did2s::event_study(
data = df |> mutate(g = ifelse(g == Inf, NA, g)) |> as.data.frame(),
gname = "g",
idname = "unit",
tname = "year",
yname = "dep_var",
estimator = "all"
)
#> Note these estimators rely on different underlying assumptions. See Table 2 of `https://arxiv.org/abs/2109.05913` for an overview.
#> Estimating TWFE Model
#> Estimating using Gardner (2021)
#> Estimating using Callaway and Sant'Anna (2020)
#> Estimating using Sun and Abraham (2020)
#> Estimating using Borusyak, Jaravel, Spiess (2021)
#> Estimating using Roth and Sant'Anna (2021)did2s::plot_event_study(multiple_ests)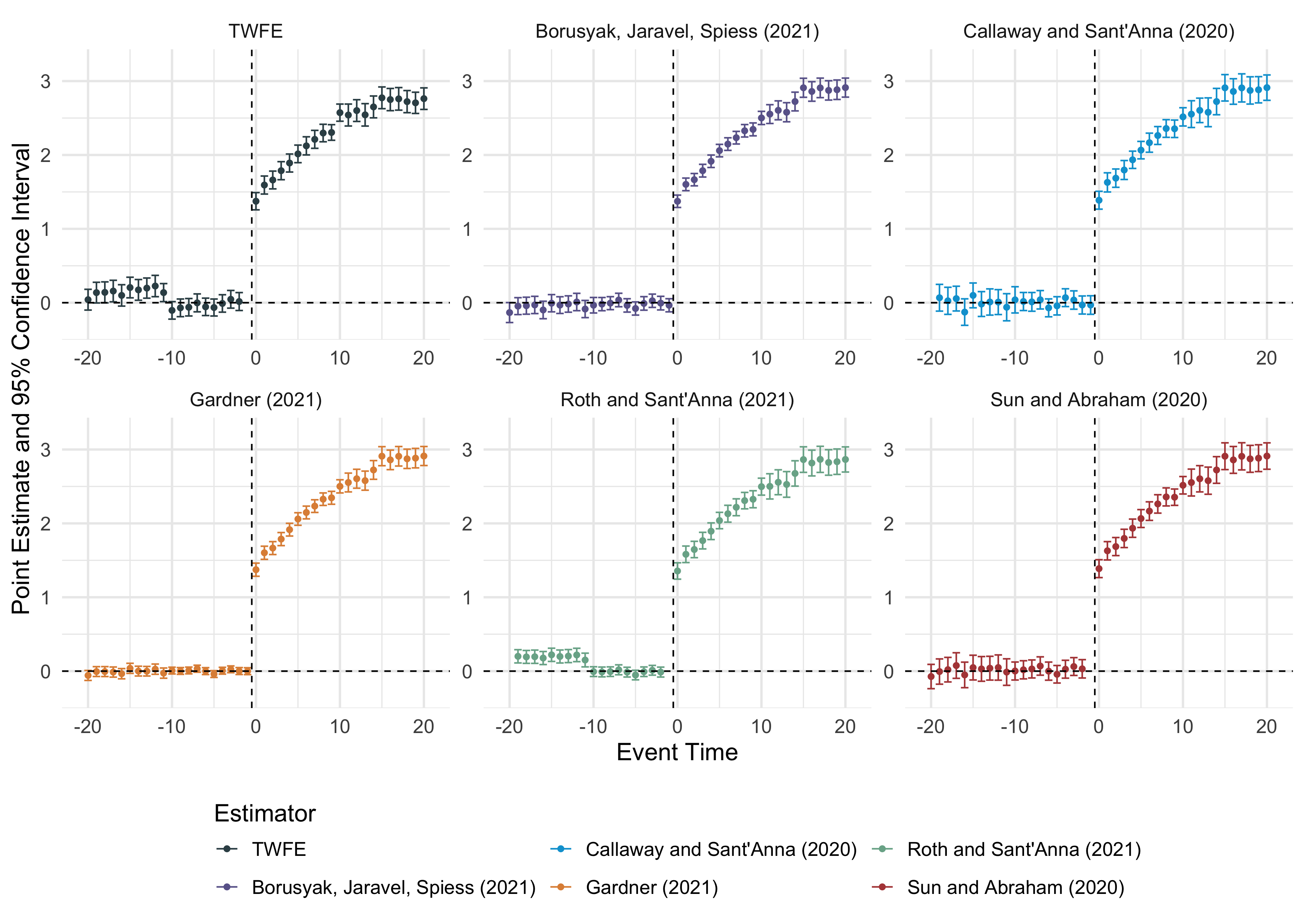
Multiple event-study estimators
If you use this package to produce scientific or commercial publications, please cite according to:
citation(package = "did2s")
#> To cite did2s in publications use:
#>
#> Butts & Gardner, "The R Journal: did2s: Two-Stage
#> Difference-in-Differences", The R Journal, 2022
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {did2s: Two-Stage Difference-in-Differences Following Gardner (2021)},
#> author = {Kyle Butts and John Gardner},
#> year = {2021},
#> url = {https://journal.r-project.org/articles/RJ-2022-048/},
#> }These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.