The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This package implement the clustering algorithm described by Alex Rodriguez and Alessandro Laio (2014). It provides the user with tools for generating the initial rho and delta values for each observation as well as using these to assign observations to clusters. This is done in two passes so the user is free to reassign observations to clusters using a new set of rho and delta thresholds, without needing to recalculate everything.
Two types of plots are supported by this package, and both mimics the
types of plots used in the publication for the algorithm. The standard
plot function produces a decision plot, with optional colouring of
cluster peaks if these are assigned. Furthermore plotMDS()
performs a multidimensional scaling of the distance matrix and plots
this as a scatterplot. If clusters are assigned observations are
coloured according to their assignment.
The two main functions for this package are
densityClust() and findClusters(). The former
takes a distance matrix and optionally a distance cutoff and calculates
rho and delta for each observation. The latter takes the output of
densityClust() and make cluster assignment for each
observation based on a user defined rho and delta threshold. If the
thresholds are not specified the user is able to supply them
interactively by clicking on a decision plot.
library(densityClust)
irisDist <- dist(iris[,1:4])
irisClust <- densityClust(irisDist, gaussian=TRUE)
#> Distance cutoff calculated to 0.2767655
plot(irisClust) # Inspect clustering attributes to define thresholds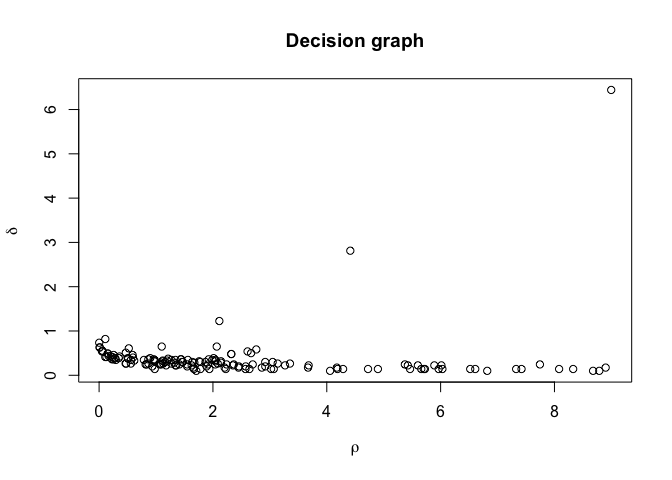
irisClust <- findClusters(irisClust, rho=2, delta=2)
plotMDS(irisClust)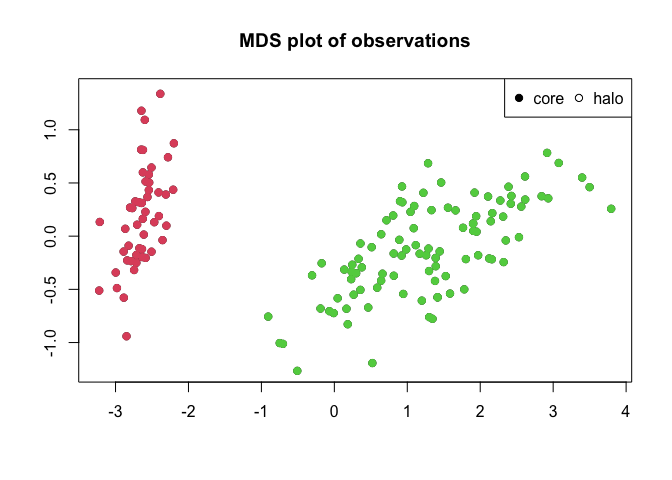
split(iris[,5], irisClust$clusters)
#> $`1`
#> [1] setosa setosa setosa setosa setosa setosa setosa setosa setosa setosa
#> [11] setosa setosa setosa setosa setosa setosa setosa setosa setosa setosa
#> [21] setosa setosa setosa setosa setosa setosa setosa setosa setosa setosa
#> [31] setosa setosa setosa setosa setosa setosa setosa setosa setosa setosa
#> [41] setosa setosa setosa setosa setosa setosa setosa setosa setosa setosa
#> Levels: setosa versicolor virginica
#>
#> $`2`
#> [1] versicolor versicolor versicolor versicolor versicolor versicolor
#> [7] versicolor versicolor versicolor versicolor versicolor versicolor
#> [13] versicolor versicolor versicolor versicolor versicolor versicolor
#> [19] versicolor versicolor versicolor versicolor versicolor versicolor
#> [25] versicolor versicolor versicolor versicolor versicolor versicolor
#> [31] versicolor versicolor versicolor versicolor versicolor versicolor
#> [37] versicolor versicolor versicolor versicolor versicolor versicolor
#> [43] versicolor versicolor versicolor versicolor versicolor versicolor
#> [49] versicolor versicolor virginica virginica virginica virginica
#> [55] virginica virginica virginica virginica virginica virginica
#> [61] virginica virginica virginica virginica virginica virginica
#> [67] virginica virginica virginica virginica virginica virginica
#> [73] virginica virginica virginica virginica virginica virginica
#> [79] virginica virginica virginica virginica virginica virginica
#> [85] virginica virginica virginica virginica virginica virginica
#> [91] virginica virginica virginica virginica virginica virginica
#> [97] virginica virginica virginica virginica
#> Levels: setosa versicolor virginicaNote that while the iris dataset contains information on three different species of iris, only two clusters are detected by the algorithm. This is because two of the species (versicolor and virginica) are not clearly seperated by their data.
Rodriguez, A., & Laio, A. (2014). Clustering by fast search and find of density peaks. Science, 344(6191), 1492-1496. https://doi.org/10.1126/science.1242072
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.