The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

The goal of denguedatahub is to provide the research
community with a unified dataset by collecting worldwide dengue-related
data, merged with exogenous variables helpful for a better understanding
of the spread of dengue and the reproducibility of research.

Check out the website at https://denguedatahub.netlify.app/
You can install the development version of denguedatahub from GitHub with:
install.packages("denguedatahub")# install.packages("devtools")
devtools::install_github("thiyangt/denguedatahub")This is a basic example which shows you how to solve a common problem:
library(tsibble)
#> Registered S3 method overwritten by 'tsibble':
#> method from
#> as_tibble.grouped_df dplyr
#>
#> Attaching package: 'tsibble'
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, union
library(denguedatahub)
head(level_of_risk)
#> # A tibble: 6 × 4
#> country level_of_risk region last_accessed
#> <chr> <chr> <chr> <date>
#> 1 Angola Sporadic/Uncertain Africa 2023-01-16
#> 2 Benin Sporadic/Uncertain Africa 2023-01-16
#> 3 Burkina Faso Frequent/Continuous Africa 2023-01-16
#> 4 Burundi Sporadic/Uncertain Africa 2023-01-16
#> 5 Cameroon Sporadic/Uncertain Africa 2023-01-16
#> 6 Cape Verde Sporadic/Uncertain Africa 2023-01-16head(srilanka_weekly_data)
#> # A tibble: 6 × 6
#> year week start.date end.date district cases
#> <dbl> <dbl> <chr> <chr> <chr> <dbl>
#> 1 2006 52 12/23/2006 12/29/2006 Colombo 71
#> 2 2006 52 12/23/2006 12/29/2006 Gampaha 12
#> 3 2006 52 12/23/2006 12/29/2006 Kalutara 12
#> 4 2006 52 12/23/2006 12/29/2006 Kandy 20
#> 5 2006 52 12/23/2006 12/29/2006 Matale 4
#> 6 2006 52 12/23/2006 12/29/2006 NuwaraEliya 1library(ggplot2)
library(viridis)
#> Loading required package: viridisLite
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
country_weekly <- srilanka_weekly_data |>
group_by(year, week, start.date) %>%
summarise(total_cases = sum(cases, na.rm = TRUE), .groups = 'drop') |>
arrange(start.date)
country_weekly <- country_weekly |>
mutate(
yearweek = yearweek(start.date)) |>
distinct(yearweek, .keep_all = TRUE)
country_weekly_tsibble <- country_weekly |>
as_tsibble(index = yearweek)
p1 <- ggplot(country_weekly_tsibble, aes(x = yearweek, y = total_cases)) +
geom_line() +
scale_x_yearweek(date_breaks = "1 year", date_labels = "%Y") +
labs(
x = "Year",
y = "Weekly Dengue Cases"
) +
theme_minimal() +
theme(
axis.text.x = element_text(angle = 45, hjust = 1),
plot.title = element_text(face = "bold")
)
p1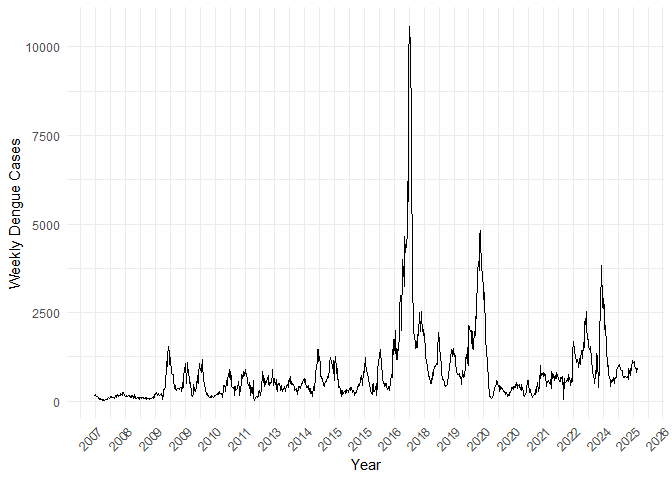
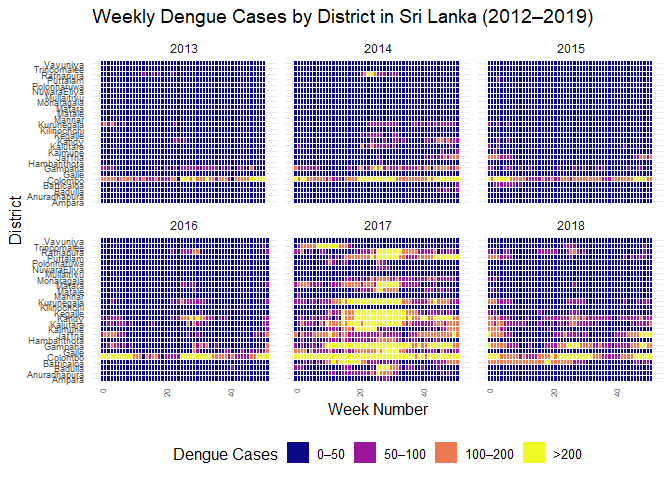
ggplot(
filter(srilanka_weekly_data, year < 2019 & year > 2012),
aes(
x = week,
y = district,
fill = cut(
cases,
breaks = c(0, 50, 100, 200, Inf),
labels = c("0–50", "50–100", "100–200", ">200"),
include.lowest = TRUE,
right = FALSE
)
)
) +
geom_tile(color = "white") +
scale_fill_viridis_d(
option = "C",
name = "Dengue Cases"
) +
facet_wrap(~year, ncol = 3) +
labs(
title = "Weekly Dengue Cases by District in Sri Lanka (2012–2019)",
x = "Week Number",
y = "District"
) +
theme_minimal(base_size = 12) +
theme(
axis.text.x = element_text(angle = 90, hjust = 1, size = 6),
axis.text.y = element_text(size = 7),
legend.position = "bottom",
strip.text = element_text(size = 9)
)
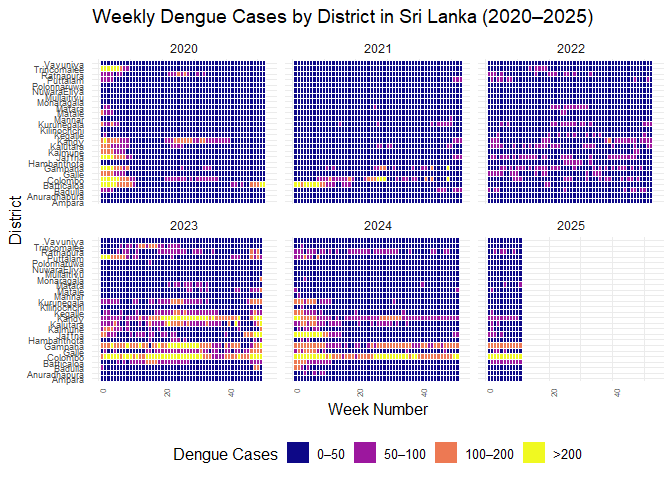
ggplot(
filter(srilanka_weekly_data, year > 2019),
aes(
x = week,
y = district,
fill = cut(
cases,
breaks = c(0, 50, 100, 200, Inf),
labels = c("0–50", "50–100", "100–200", ">200"),
include.lowest = TRUE,
right = FALSE
)
)
) +
geom_tile(color = "white") +
scale_fill_viridis_d(
option = "C",
name = "Dengue Cases"
) +
facet_wrap(~year, ncol = 3) +
labs(
title = "Weekly Dengue Cases by District in Sri Lanka (2020–2025)",
x = "Week Number",
y = "District"
) +
theme_minimal(base_size = 12) +
theme(
axis.text.x = element_text(angle = 90, hjust = 1, size = 6),
axis.text.y = element_text(size = 7),
legend.position = "bottom",
strip.text = element_text(size = 9)
)
library(tidyverse)
world_annual |>
filter(region=="Afghanistan") |>
head()
#> long lat group order region subregion code year incidence
#> 1 74.89131 37.23164 2 12 Afghanistan <NA> AFG 1990 23371
#> 2 74.89131 37.23164 2 12 Afghanistan <NA> AFG 1991 25794
#> 3 74.89131 37.23164 2 12 Afghanistan <NA> AFG 1992 29766
#> 4 74.89131 37.23164 2 12 Afghanistan <NA> AFG 1993 32711
#> 5 74.89131 37.23164 2 12 Afghanistan <NA> AFG 1994 34268
#> 6 74.89131 37.23164 2 12 Afghanistan <NA> AFG 1995 35823
#> dengue.present
#> 1 yes
#> 2 yes
#> 3 yes
#> 4 yes
#> 5 yes
#> 6 yesThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.