
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

deepredeff is a package to predict effector protein given amino acid sequences. This tool can be used to predict effectors from three different taxa, which are oomycete, fungi, and bacteria.
You can install the released version of deepredeff from
CRAN with:
install.packages("deepredeff")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("ruthkr/deepredeff")The deepredeff package uses TensorFlow. If you already
have TensorFlow 2.0.0 or later in your system, then you can specify the
environment where TensorFlow is installed using
reticulate::use_condaenv(). Otherwise, you can install
TensorFlow, by using the install_tensorflow() function as
follows:
library(deepredeff)
install_tensorflow()Note that this only needs to be run once, the first
time you use deepredeff.
To use deepredeff, you can read the documentation on the
following topics:
This is a basic example which shows you how to predict effector sequences if you have a FASTA file:
# Load the package
library(deepredeff)
# Define the fasta path from the sample data
bacteria_fasta_path <- system.file(
"extdata/example", "bacteria_sample.fasta",
package = "deepredeff"
)
# Predict the effector candidate using bacteria model
pred_result <- predict_effector(
input = bacteria_fasta_path,
taxon = "bacteria"
)
#> Loaded models successfully!
#> Model used for taxon bacteria: ensemble_weighted.# View results
pred_result| name | sequence | s_score | prediction |
|---|---|---|---|
| tr⎮A0A0N8SZV2⎮A0A0N8SZV2_PSESY Type III secretion system effector HopAI1 OS=Pseudomonas syringae pv. syringae OX=321 GN=ALO45_04155 PE=4 SV=1 | MPINRPAFNLKLNTAIAQPTLKKDA | 0.9483424 | effector |
| tr⎮A5CLR7⎮A5CLR7_CLAM3 Pat-1 protein OS=Clavibacter michiganensis subsp. michiganensis (strain NCPPB 382) OX=443906 GN=pat-1 PE=4 SV=1 | MQFMSRINRILFVAVVSLLSVLGCC | 0.0798178 | non-effector |
| sp⎮B2SU53⎮PTHX1_XANOP TAL effector protein PthXo1 OS=Xanthomonas oryzae pv. oryzae (strain PXO99A) OX=360094 GN=pthXo1 PE=1 SV=2 | MDPIRSRTPSPARELLPGPQPDRVQ | 0.9943361 | effector |
| tr⎮C0SPN9⎮C0SPN9_RALSL Uncharacterized protein RSc2139 OS=Ralstonia solanacearum OX=305 GN=RSc2139 PE=4 SV=1 | MSIGRSKSVAGASASHALASGENGS | 0.8418444 | effector |
| tr⎮D2Z000⎮D2Z000_RALSL Type III effector protein OS=Ralstonia solanacearum OX=305 GN=rip61 PE=4 SV=1 | MPPPIRNARTTPPSFDPSAAGDDLR | 0.9953785 | effector |
| tr⎮Q8XX20⎮Q8XX20_RALSO Putative multicopper oxidase, type 3 signal peptide protein OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=RSc2298 PE=4 SV=1 | MSHMTFNTWKAGLWRLAAAAVLSLL | 0.0645516 | non-effector |
| tr⎮Q87UH8⎮Q87UH8_PSESM Taurine ABC transporter, periplasmic taurine-binding protein OS=Pseudomonas syringae pv. tomato (strain ATCC BAA-871 / DC3000) OX=223283 GN=tauA PE=4 SV=1 | MKLHFSLRLLTALSLTGATFLAQAA | 0.0492858 | non-effector |
| tr⎮Q4ZTI0⎮Q4ZTI0_PSEU2 Amino acid ABC transporter substrate-binding protein, PAAT family OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=Psyr_2503 PE=4 SV=1 | MHRGPSFVKACAFVLSASFMLANTV | 0.3061618 | non-effector |
| tr⎮Q4ZR15⎮Q4ZR15_PSEU2 Sensor protein OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=Psyr_3375 PE=4 SV=1 | MRRQPSLTLRSTLAFALVAMLTVSG | 0.0722144 | non-effector |
| tr⎮D4I1R4⎮D4I1R4_ERWAC Outer-membrane lipoprotein LolB OS=Erwinia amylovora (strain CFBP1430) OX=665029 GN=lolB PE=3 SV=1 | MLSSNRRLLRLLPLASLLLTACGLH | 0.0489914 | non-effector |
After getting the prediction results, you can plot the probability distribution of the results as follows:
plot(pred_result)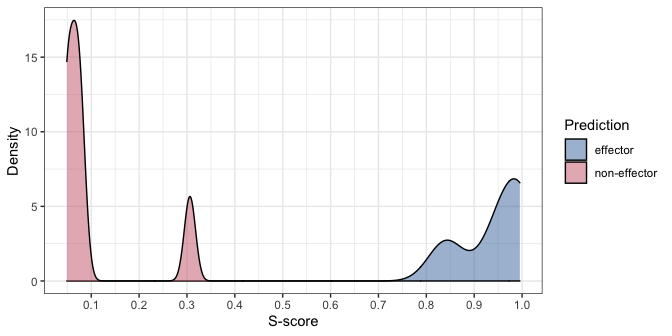
More examples with different input formats are available on functions documentations and vignettes, please refer to the documentation.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.