

The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
ddecompose:
Detailed Distributional Decompositions in R
ddecompose:
Detailed Distributional Decompositions in RThe R package ddecompose implements the Oaxaca (1973)-Blinder (1973) decomposition method and generalizations of it that decompose differences in distributional statistics beyond the mean.
ob_decompose() decomposes differences in the mean
outcome between two groups into one part explained by different
covariates (composition effect) and into another part due to differences
in linear regression coefficients linking the covariates to the outcome
variable (structure effect). The function further divides the two
effects into the contribution of each covariate and allows for weighted
‘doubly robust’ decompositions. For distributional statistics beyond the
mean, the function performs the RIF decomposition of Firpo, Fortin, and
Lemieux (2018).
dfl_decompose() divides differences in distributional
statistics into a composition effect and a structure effect using
inverse probability weighting as proposed by DiNardo, Fortin, and Lemieux
(1996). The function also allows to sequentially decompose the
composition effect into the contribution of single covariates.
The package contains generic summary, print and plot functions for the results and computes standard errors. This documentation provides a brief overview of the functions implemented in the package. For a more detailed discussion of decomposition methods, including their respective assumptions and limitations, refer to Fortin, Lemieux, and Firpo (2011) and Firpo et al. (2018).
In contrast to the existing oaxaca (Hlavac, 2022), ddecompose is not limited to decomposition of mean differences and offers ‘doubly robust’ decompositions. The package also complements Counterfactual (Chen et al., 2017) that models conditional distribution functions instead of using inverse probability weighting to perform aggregate decompositions beyond the mean.
You can hopefully soon install the CRAN version of
ddecompose
install.packages("ddecompose")Until now, refer to the latest development version from GitHub:
install.packages("devtools")
devtools::install_github("samumei/ddecompose")The original decomposition method introduced by Oaxaca (1973) and Blinder (1973) divides the difference in the mean of an outcome variable (e.g., hourly wages) between two groups \(g = 0, 1\) into a part explained by differences in the mean of the covariates (e.g., educational level or experience) and into another part due to different linear regression coefficients (e.g., returns to education) that link the covariates to the outcome variable.
The method linearly models the relationship between the outcome \(Y\) and covariates \(X\) \[Y_{g,i} = \beta_{g,0} + \sum^K_{k=1}X_{k,i}\beta_{g,k} + \varepsilon_{g,i},\] where \(\beta_{g,0}\) is the intercept and \(\beta_{g,k}\) are the slope coefficients of covariates \(k = 1,\ldots, K\). Moreover, it is assumed that the error term \(\varepsilon\) is conditionally independent of \(X\), i.e., \(E( \varepsilon_{g,i} | X_{1,i}, \ldots ,X_{k,i}) = 0\), and that there is an overlap in observable characteristics across groups (‘common support’).
The coefficients are estimated with OLS. Together with the sample
means of the covariates, one can derive a counterfactual mean
\[\overline Y_C = \widehat \beta_{0,0} +
\sum^K_{k=1}\overline X_{1,k} \widehat \beta_{0,k}\] that would
be observed if group \(1\) had the same
coefficients like group \(0\). By
adding and subtracting the counterfactual, the observed difference \[\widehat\Delta^\mu_O = (\overline Y_1 - \overline
Y_C) + (\overline Y_C - \overline Y_0) = \widehat\Delta^\mu_S +
\widehat\Delta^\mu_C, \]
is divided into the aggregate structure effect \[\widehat\Delta^\mu_S = (\widehat \beta_{1,0} - \widehat \beta_{0,0}) + \sum^K_{k=1}\overline X_{1,k}(\widehat \beta_{1,k} - \widehat \beta_{0,k}),\]
that captures outcome differences due to different coefficients, and the composition effect \[\widehat\Delta^\mu_X = \sum^K_{k=1} (\overline X_{1,k} - \overline X_{0,k})\widehat \beta_{0,k}, \]
which accounts for mean differences of the covariates. Note that we could also combine the coefficients from group 1 with the covariates of group 0 to define a counterfactual. Such a change in the “reference” group generally leads to different results.
\(\widehat\Delta^\mu_S\) and \(\widehat\Delta^\mu_X\) denote the aggregate composition terms. Since the method hinges on an additive linear model, the terms can be further divided into the contribution of each covariate \(k\) in a detailed decomposition. For instance, the contribution of covariate \(k = 1\) to the structure effect is \(\overline X_{1,1}(\widehat \beta_{1,1} - \widehat \beta_{0,1})\), while \((\overline X_{1,1} - \overline X_{0,1})\widehat \beta_{0,1}\) is the covariate’s contribution to the composition effect.
The Oaxaca-Blinder decomposition is limited to the mean. Moreover, the decomposition terms are biased if the expectation of the outcome conditional on the covariates is not linear (see Barsky et al., 2002). DiNardo, Fortin, and Lemieux (1996), DFL hereafter, propose an alternative approach that overcomes both shortcomings. Instead of modeling the conditional mean, the method uses inverse probability weighting to estimate a counterfactual outcome distribution that combines the conditional outcome distribution of one group and the covariates distribution of the other group. For instance, if we are interested in the outcomes of group 0 with covariates of group 1, we would reweight the outcome distribution of group 0 such that its covariates distribution matches that of group 1 \[F_{Y_C}(y) = \int F_{Y_0}(y|x)dF_{X_1} (x)= \int F_{Y_0}(y|x)\Psi_X(x)dF_{X_0}(x).\]
By applying Bayes’ rule, the reweighting factor, \[\Psi_X(x) = \frac{dF_{X_1}(x)}{dF_{X_0}(x)} = \frac{P(g=0)P(g=1|x)}{P(g=1)P(g=0|x)},\]
can be expressed in terms of \(P(g)\) and \(P(g|x)\), the (conditional) probabilities of belonging to group \(g\). This allows us to estimate the reweighting factor using sample probabilities of each group and fitting conditional probability models (e.g., logit) in the joint sample. The estimated factors are then used to estimate weighted distributional statistics of interest (e.g., mean, quantiles or Gini coefficient) in the reference sample – group 0 in the present example. The resulting counterfactual distributional statistic, \(\widehat\nu_C=\widehat\nu(F_{Y_C})\), is then contrasted with the observed difference \[\widehat\Delta_O^{\nu} = (\widehat\nu_1 - \widehat\nu_C) + (\widehat\nu_C - \widehat\nu_0) = \widehat\Delta_S^\nu + \widehat\Delta_X^\nu,\]
which yields again an aggregate structure effect and aggregate composition effect.
The two decomposition terms account for the contribution of the covariates and of the conditional outcome distribution, respectively, assuming common support and ignorability. The latter condition asserts that the distribution of unobserved covariates \(\varepsilon\) conditional on observed covariates \(X\) is independent of group \(g\).
In contrast to the Oaxaca-Blinder decomposition, where the contributions of each covariate simply add up, detailed decompositions are not straightforward in the reweighting framework. However, DFL show that we can sequentially alter the covariates distributions to decompose the composition effect into the contribution of single covariates. For instance, assume we want to distinguish the effect of covariate \(X_1\) (e.g., union status) from that of covariate \(X_2\) (e.g., industry). We begin again with the counterfactual distribution based on the conditional outcome distribution of group 0 and the covariates distribution of group 1 \[F_{Y_{C}}(y) = \iint F_{Y_0}(y|x_1,x_2)dF_{X_{1,1}}(x_1|x_2)dF_{X_{1,2}}(x_2)\]
and introduce a second counterfactual where we combine the conditional outcome distribution of group 0 as well as the conditional covariate distribution of \(X_1\) given \(X_2\) (e.g., union coverage by industry) of group 0 with the covariates distribution \(X_2\) (e.g., employment by industry) of group 1 \[F_{Y_{C,X_2}}(y) = \iint F_{Y_0}(y|x_1,x_2)dF_{X_{0,1}}(x_1|x_2)dF_{X_{1,2}}(x_2) \]
which can be expressed as the outcome distribution 0 \[F_{Y_{C,X_2}}(y) = \iint F_{Y_0}(y|x_1,x_2)dF_{X_{0,1}}(x_1|x_2)\Psi_{X_2}(x_2)dF_{X_{0,2}}(x_2),\] reweighted by the factor \[\Psi_{X_2}(x_2) = \frac{dF_{X_{1,2}}(x_2)}{dF_{X_{0,2}}(x_2)} = \frac{P(g=0)P(g=1|x_2)}{P(g=1)P(g=0|x_2)}.\]
Using the distributional statistics of the additional counterfactual, we can divide the aggregate decomposition effect into the contribution of each covariate \[\widehat \Delta_X^{\nu} = (\widehat \nu_C - \widehat \nu_{C,X_2}) + (\widehat \nu_{C,X_2} - \widehat \nu_0) = \widehat \Delta_{X_1}^\nu + \widehat \Delta_{X_2}^\nu.\]
Sequential decompositions are path dependent because the detailed composition effects attributed to single covariates depend on the order in which we include the variables into the sequence. For instance, it matters whether we reweight union coverage by industry \(F(x_1|x_2)\) or the industry employment given union coverage \(F(x_2|x_1)\).
Moreover, we get different results if we do not combine the marginal covariate distribution \(X_2\) (e.g., industry employment) of group 1 with the conditional distribution of \(X_1\) given \(X_2\) (e.g., union density by industry) of group 0 but rather, combine the marginal of group 0 with the conditional distribution of group 1 to derive the counterfactual, e.g., \[F_{Y_{C,X_1}}(y) = \iint F_{Y_0}(y|x_1,x_2)dF_{X_{1,1}}(x_1|x_2)dF_{X_0,2}(x_2).\] where we would reweight group 0 with a slightly different factor \[\Psi_{X_1}(x_1,x_2) = \frac{dF_{X_{1,1}}(x_1|x_2)}{dF_{X_{0,1}}(x_1|x_2)} = \frac{P(g=0|x_2)P(g=1|x_1,x_2)}{P(g=1|x_2)P(g=0|x_1,x_2)}.\]
A robust and path independent alternative for detailed decompositions at the mean is to combine DFL reweighting with the linear Oaxaca-Blinder method (see Fortin et al., 2011: 48-51). This approach has the valuable side effect of accounting for potential errors introduced by an incomplete inverse probability weighting and the linear model specification, respectively.
The estimation proceeds in two steps. First, the reweighting function \(\widehat\Psi_X(x)\), which matches the characteristics of group \(0\) (if group 0 is the reference group) to those of group \(1\), is derived. Second, the linear model and the covariate means are estimated in the two observed samples as well as in the reweighted sample \(0\). We denote the estimates of the reweighted sample as \(\widehat \beta_{C,0}\), \(\widehat \beta_{C,k}\), and \(\overline X_{C,k}\).
The mean of the reweighted sample builds the main counterfactual to derive the aggregate structure and composition effects, respectively. The detailed decomposition terms are also evaluated with respect to the statistics of the reweighted sample, i.e.,
\[\widehat\Delta^\mu_{O,R} = (\widehat \beta_{1,0} - \widehat \beta_{C,0}) + \sum^K_{k=1} (\overline X_{1,k}\widehat \beta_{1,k} - \overline X_{C,k}\widehat \beta_{C,k}) + \sum^K_{k=1} (\overline X_{C,k}\beta_{C,k} - \overline X_{0,k}\widehat \beta_{0,k}) = \widehat\Delta^\mu_{S,R} + \widehat\Delta^\mu_{X,R}.\] These decomposition terms can be further decomposed into a structure and into a composition effect, respectively. This separates the errors from reweighting \(\widehat\Delta^\mu_{S,e}\) and the linear specification \(\widehat\Delta^\mu_{X,e}\), respectively, and yields a “pure” composition effect \(\widehat\Delta^\mu_{X,p}\) and a “pure” structure effect \(\widehat\Delta^\mu_{S,p}\) for each covariate.
Specifically, the structure effect can be written as
\[\widehat\Delta^\mu_{S,R} = (\widehat \beta_{1,0} - \widehat \beta_{C,0}) + \sum^K_{k=1}\overline X_{1,k}(\widehat \beta_{1,k} - \widehat \beta_{C,k}) + \sum^K_{k=1} (\overline X_{1,k} - \overline X_{C,k})\widehat \beta_{C,k} = \widehat\Delta^\mu_{S,p} + \widehat\Delta^\mu_{S,e}.\]
By comparing \(\beta_{1,k}\) to the coefficients of the reweighted group \(0\), \(\widehat \beta_{C,k}\), a “pure” structure effect adjusted for the reweighting error, \(\widehat\Delta^\mu_{S,p}\), can be identified. Moreover, the reweighting error, \(\widehat\Delta^\mu_{S,e}\), indicates how accurately the reweighting function \(\widehat\Psi(X_i)\) reweights the covariates distribution of group \(0\) to that of group \(1\). If the composition of group \(1\) is equal to the reweighted group \(0\), the reweighting error is zero.
Similarly, the additional decomposition of the composition effect reads as
\[\widehat\Delta^\mu_{X,R} = \sum^K_{k=1} (\overline X_{C,k} - \overline X_{0,k})\widehat \beta_{0,k} + (\widehat \beta_{C,0} - \widehat \beta_{0,0}) + \sum^K_{k=1}\overline X_{C,k}(\widehat \beta_{C,k} - \widehat \beta_{0,k}) = \widehat\Delta^\mu_{X,p} + \widehat\Delta^\mu_{X,e}.\]
The specification error, \(\widehat\Delta^\mu_{X,e}\), measures the extent to which the estimated coefficients change due to a different distribution of covariates if the (wage) structure remains the same. Thereby, the “pure” effect, \(\overline X_{C,k} - \overline X_{0,k}\), for each covariate can be estimated, summing to the aggregate pure structure effect, \(\widehat\Delta^\mu_{X,p}\). If the model is truly linear, i.e., correctly specified, \(\beta_C\) of the reweighted group \(0\) will be identical to \(\beta_0\) and the specification error will be zero (Fortin et al. 2011, pp. 49-50).
The reweighted OB decomposition is “doubly robust” as it yields consistent estimates even if either the linear model or the reweighting estimator is misspecified. In contrast to the simple reweighting or linear approach it does not hinge on a single correctly specified model. While the reweighted OB decomposition is doubly robust and path independent, it is limited to the mean.
A path independent method that goes beyond the mean is the RIF
decomposition of Firpo, Fortin, and Lemieux (2018). The approach
approximates the expected value of the ‘recentered influence function’
(RIF) of the distributional statistic (e.g., quantile, variance, or Gini
coefficient) of an outcome variable conditional on covariates with
linear regressions. RIF regression coefficients are consistent estimates
of the marginal effect of a small change in the expected value of a
covariate on the distributional statistics of an outcome variable (see
Firpo
et al., 2009a and the documentation of the companion package rifreg). Thus,
they can be used to decompose between-group difference in distributional
statistics. Firpo et al. (2018) combine the RIF regressions again with
the reweighting estimator to avoid specification errors.
First, the approach computes the recentered influence function (RIF) of the outcome variable \(Y\) and a distributional statistic of interest \(\nu\). Then, an OLS regression of the transformed outcome variable \(Y\) is run on the explanatory variables \(X\). Thereafter, the decomposition method is analogous to the original OB method:
\[\widehat\Delta^\nu_{O,R} = \widehat\Delta^\nu_{S,p} + \widehat\Delta^\nu_{S,e} + \widehat\Delta^\nu_{X,p} + \widehat\Delta^\nu_{X,e}.\] With \(\widehat\Delta^\nu_{S,p}\) and \(\widehat\Delta^\nu_{S,e}\) estimating the pure structure effect and the reweighting error as
\[\widehat\Delta^\nu_{S,R} = \widehat\Delta^\nu_{S,p} + \widehat\Delta^\nu_{S,e} = (\widehat \beta_{1,0} - \widehat \beta_{C,0}) + \sum^K_{k=1}\overline X_{1,k}(\widehat \beta_{1,k} - \widehat \beta_{C,k}) + \sum^K_{k=1} (\overline X_{1,k} - \overline X_{C,k})\widehat \beta_{C,k}\]
and \(\widehat\Delta^\nu_{X,p}\) and \(\widehat\Delta^\nu_{X,e}\) estimating the pure coefficient effect and the specification error:
\[\widehat\Delta^\nu_{X,R} = \widehat\Delta^\nu_{X,p} + \widehat\Delta^\nu_{X,e} = \sum^K_{k=1} (\overline X_{C,k} - \overline X_{0,k})\widehat \beta_{0,k} + (\widehat \beta_{C,0} - \widehat \beta_{0,0}) + \sum^K_{k=1}\overline X_{C,k}(\widehat \beta_{C,k} - \widehat \beta_{0,k})\] with the RIF regression coefficients \(\widehat \beta\) and the covariate means \(\overline X\) of groups \(0\), \(1\), and \(C\), the reweighted reference group 0, respectively.
Again, the specification error increases if the conditional expectation of the RIF is not well approximated by the linear model. Moreover, as the RIF regression coefficients capture the marginal effects of small location shifts in the covariates distribution on the distributional statistic of the outcome variable, the specification error can be large if the location shifts are substantial or if the between-group differences in the covariates distribution relate to higher moments or the dependence structure among the covariates (see also Rothe, 2012: 16-19 or Rothe, 2015: 328).
Analytical standard errors are straightforward for the Oaxaca-Blinder
decomposition under the assumption of independence between the groups
(see Jann, 2005 and
2008).
Firpo
(2007) and Firpo and
Pinto (2016) develop asymptotic theory for the reweighting
estimator. Firpo,
Fortin, and Lemieux (2009b) and Firpo et al. (2018) do the same for
the RIF estimator and for the RIF decomposition terms, respectively. The
authors propose bootstrapping the standard errors. In
ddecompose, the standard errors can be bootstrapped in both
ob_decompose() and dfl_decompose(). For the
linear Oaxaca-Blinder decomposition, analytical standard errors are also
available.
The following examples illustrate the operation of the main
decomposition functions in ddecompose. We use a sample from
the National Longitudinal Survey (NLSY) 79 containing wage data from the
year 2000 for workers who aged 35 to 43. The data are from O’Neill and
O’Neill (2006) and were used to illustrate the Oaxaca-Blinder mean
decomposition in Fortin, Lemieux, and Firpo (2011). The data contains
2655 male and 2654 female observations, respectively.
library(ddecompose)
data("nlys00")We decompose the gender wage gap into composition and structure effect, using the Oaxaca-Blinder decomposition without reweighting. We specify a wage regression model and then run the estimation.
model <- log(wage) ~ age + region + education +
years_worked_civilian + years_worked_military +
part_time + family_responsibility + industry
gender_gap_decomposition <- ob_decompose(
formula = model,
data = nlys00,
group = female
)By default, the function subtracts the mean of the lower rank group
from the mean of the higher rank group to calculate the overall
difference, and uses the coefficients of the lower ranked group to
calculate the counterfactual. In the present example, yes
is the reference level and the lower ranked value of the factor variable
female. Thus, the female mean is subtracted from the male
mean and the female coefficients are used to estimate the counterfactual
mean.
levels(nlys00$female)
#> [1] "yes" "no"If we want to change the reference group, i.e., use male coefficients
to estimate the counterfactual, we have to change the parameter
reference_0 = FALSE so that the higher ranked group is used
as reference. To change the direction of the subtraction, you can change
the parameter subtract_1_from_0.
gender_gap_decomposition <- ob_decompose(
formula = model,
data = nlys00,
group = female,
reference_0 = FALSE,
subtract_1_from_0 = TRUE
)With summary(), we can display the decomposition formula
and the estimation results. For Oaxaca-Blinder decompositions, the
function displays standard errors by default, assuming independence
between the groups and homoscedasticity.
summary(gender_gap_decomposition)
#>
#>
#> Oaxaca-Blinder decomposition of mean difference
#> between female == 'no' (group 1) and female == 'yes' (group 0).
#> The reference group is 'no'.
#>
#> Group 0: female == 'yes' (2654 observations)
#> Group 1: female == 'no' (2655 observations)
#>
#> Composition Effect: (X0 - X1) * b1
#> Structure Effect: X0 * (b0 - b1)
#>
#> Aggregate decomposition:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Observed difference -0.23330029 0.01466550 -0.26204413 -0.2045564
#> Composition effect -0.20686977 0.01828421 -0.24270616 -0.1710334
#> Structure effect -0.02643052 0.01907475 -0.06381634 0.0109553
#>
#>
#> Observed difference:
#>
#> Estimate Std. Error [ 95%-CI ]
#> (Intercept) -0.09813187 0.213742888 -0.517060232 0.32079649
#> age 0.19459845 0.231908532 -0.259933920 0.64913082
#> region -0.03970035 0.027067240 -0.092751163 0.01335047
#> education -0.05379461 0.033738154 -0.119920179 0.01233096
#> years_worked_civilian -0.27105219 0.061730110 -0.392040980 -0.15006340
#> years_worked_military -0.01683904 0.003284003 -0.023275567 -0.01040251
#> part_time 0.01470307 0.008433550 -0.001826383 0.03123253
#> family_responsibility -0.03558673 0.011343387 -0.057819361 -0.01335410
#> industry 0.07250298 0.031630225 0.010508877 0.13449708
#>
#>
#> Structure effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> (Intercept) -0.0981318689 0.2137428884 -0.5170602321 0.320796494
#> age 0.2024835070 0.2327690880 -0.2537355222 0.658702536
#> region -0.0353034766 0.0271166306 -0.0884510959 0.017844143
#> education -0.0742506682 0.0335841725 -0.1400744368 -0.008426900
#> years_worked_civilian -0.1954027885 0.0575554072 -0.3082093137 -0.082596263
#> years_worked_military 0.0008898637 0.0008924724 -0.0008593501 0.002639078
#> part_time 0.0813405909 0.0153752265 0.0512057007 0.111475481
#> family_responsibility -0.0037184554 0.0179140863 -0.0388294193 0.031392509
#> industry 0.0956627763 0.0335458386 0.0299141407 0.161411412
#>
#>
#> Composition effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> (Intercept) 0.000000000 3.725652e-15 -7.302144e-15 7.302144e-15
#> age -0.007885058 2.292319e-03 -1.237792e-02 -3.392194e-03
#> region -0.004396870 2.357891e-03 -9.018252e-03 2.245116e-04
#> education 0.020456057 7.611010e-03 5.538752e-03 3.537336e-02
#> years_worked_civilian -0.075649400 7.599371e-03 -9.054389e-02 -6.075491e-02
#> years_worked_military -0.017728902 2.902154e-03 -2.341702e-02 -1.204079e-02
#> part_time -0.066637519 9.159341e-03 -8.458950e-02 -4.868554e-02
#> family_responsibility -0.031868276 1.096548e-02 -5.336023e-02 -1.037632e-02
#> industry -0.023159799 6.351270e-03 -3.560806e-02 -1.071154e-02ddecompose comes with a handy plotting function. To plot
the overall composition and structure effects, we need to set
detailed_effects = FALSE.
plot(gender_gap_decomposition, detailed_effects = FALSE)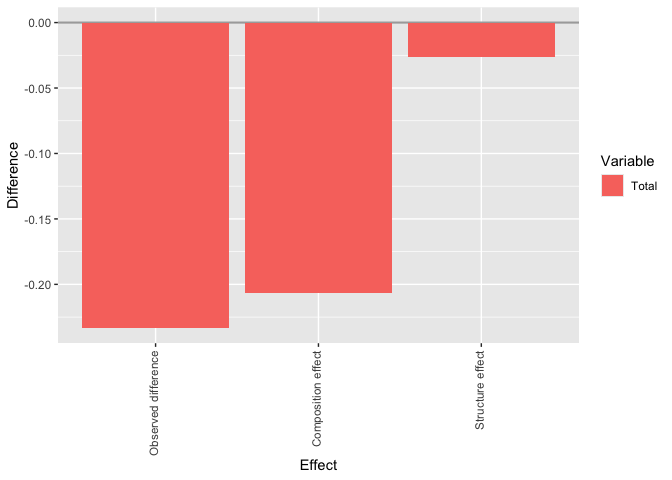
Note that detailed decompositions with factor variables depend on the
factor’s left-out reference level or base group.
ob_decompose allows to normalize factor variables with the
approach of Gardeazabal and Ugidos (2004) by setting
normalize_factors = TRUE to get detailed decompositions
that are independent of the reference level.
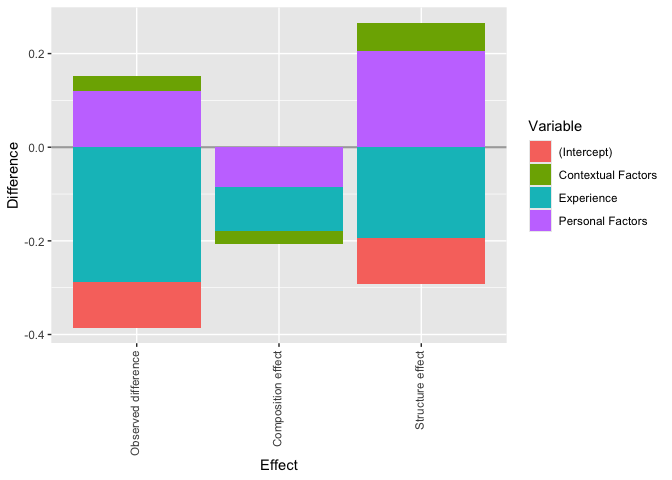
We can aggregate the detailed effects displayed in the
summary() and plot() functions. For example,
if we want to separate personal and contextual factors that explain the
wage gap, we can aggregate these variables in a list.
custom_aggregation <- list(
`Personal Factors` = c(
"age",
"education<10 yrs",
"educationHS grad (diploma)",
"educationHS grad (GED)",
"educationSome college",
"educationBA or equiv. degree",
"educationMA or equiv. degree",
"educationPh.D or prof. degree",
"part_time",
"family_responsibility"
),
`Experience` = c(
"years_worked_civilian",
"years_worked_military"
),
`Contextual Factors` = c(
"regionNorth-central",
"regionSouth",
"regionWest",
"industryManufacturing",
"industryEducation, Health, Public Admin.",
"industryOther services"
)
)
plot(gender_gap_decomposition,
custom_aggregation = custom_aggregation
)
To estimate a ‘doubly robust’ decomposition, we can add reweighting
to the decomposition. Thereby, we also estimate the specification and
reweighting errors. By default, the same covariate specification is used
for the outcome model as for the conditional probability model used to
derive the reweighting factors. However, it is advisable to add a more
flexible reweighting model, taking interactions into account. The
reweighting formula is added to the decomposition formula, separated by
the | operator.
For decomposition based on reweighting, standard errors need to be
bootstrapped. By increasing the number of cores, bootstrap
computation can be parallelized, thereby reducing computation time. By
default, 100 bootstrap replications are calculated. If
bootstrap = FALSE (default) no standard errors are
computed.
model_w_reweighting <- log(wage) ~
age + region + education +
years_worked_civilian + years_worked_military +
part_time + family_responsibility + industry |
age + region + education +
years_worked_civilian + years_worked_military +
part_time + family_responsibility + industry +
education:region + age:education
gender_gap_decomposition_w_reweighting <- ob_decompose(
formula = model_w_reweighting,
data = nlys00,
group = female,
reference_0 = FALSE,
reweighting = TRUE,
bootstrap = TRUE,
cores = 4
)The default method for fitting and predicting conditional
probabilities, used to derive the reweighting factor, is a logit model.
However, you can also use reweighting_method = "fastglm" to
fit a logit model with the fast algorithm of fastglm,
or reweighting_method = "random_forest" to estimate the
conditional probabilities with the ranger
implementation of random forest.
Setting trimming = TRUE will trim observations with
dominant reweighting factor values. By default, reweighting factor
values are trimmed according to the rule of Huber, Lechner, and Wunsch
(2013). Thereby, the trimming_threshold, i.e., the maximum
accepted relative weight of the reweighting factor value (inverse
probability weight) of a single observation, is set to
sqrt(N)/N, where N is the number of
observations in the reference group. The trimming threshold can also be
manually set to a numeric value.
If we add reweighting, the plot() and
summary() functions will also display the specification and
reweighting error.
To decompose group differences beyond the mean with
ob_decompose we use RIF regressions. In the following
examples, we will analyze the changes in wage inequality between 1983/85
and 2003/05 and assess which covariates contribute to explaining these
changes. In this example, the two groups are identified by the variable
year, the lower ranked year '1983-1985' is
used as reference group. First, we look at the changes in the variance.
Then, we decompose the wage gap at each decile. We use a subsample of
the CPS data used in the handbook chapter of Fortin, Lemieux, &
Firpo (2011).
data("men8305")
model_rifreg <- log(wage) ~ union + education + experience |
union * (education + experience) +
education * experience
# Variance
variance_decomposition <- ob_decompose(
formula = model_rifreg,
data = men8305,
group = year,
reweighting = TRUE,
rifreg_statistic = "variance",
bootstrap = TRUE,
cores = 4
)
# Deciles
deciles_decomposition <- ob_decompose(
formula = model_rifreg,
data = men8305,
group = year,
reweighting = TRUE,
rifreg_statistic = "quantiles",
rifreg_probs = c(1:9) / 10,
bootstrap = TRUE,
cores = 4
)
# Plotting the deciles
plot(deciles_decomposition)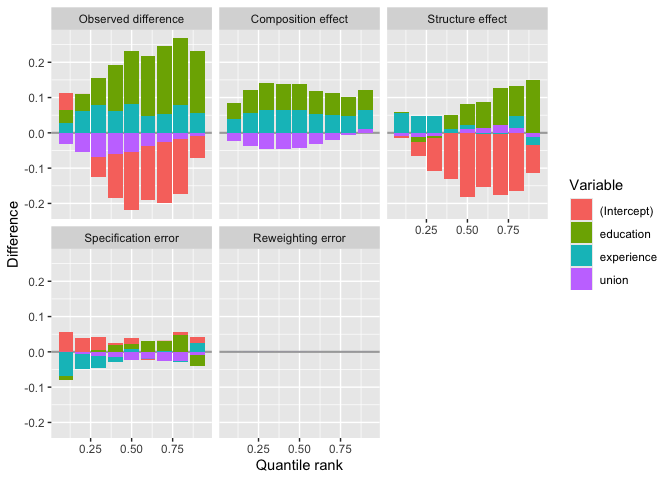
The RIF functions for the following statistics are currently
implemented: "quantiles", "mean",
"variance", "gini",
"interquantile_range", and
"interquantile_ratio". However, ob_decompose
also allows you to pass a custom RIF function for the decomposition, by
setting rifreg_statistic = "custom" and passing the custom
function to custom_rif_function. Cowell and
Flachaire (2007), Essama-Nssah
& Lambert (2012), and Rios-Avila (2020)
derive the influence functions for an array of distributional
statistics. More information about RIF regressions can be found in the
documentation of the companion package rifreg.
Custom RIF functions must specify a dep_var parameter
for the outcome variable \(Y\),
weights for potential sample weights, and
probs defining the corresponding probabilities of
quantiles. If they are not needed, they must be set to NULL
in the function definition (e.g., probs = NULL).
The following example shows how to write the RIF for the top 10 percent income share and, then, to estimate the RIF regression decomposition using this custom function. The formula for this specific RIF can be found in Essam-Nssah & Lambert (2012) or Rios-Avila (2020).
# custom RIF function for top 10% percent income share
custom_top_inc_share <- function(dep_var,
weights,
probs = NULL,
top_share = 0.1) {
top_share <- 1 - top_share
weighted_mean <- weighted.mean(
x = dep_var,
w = weights
)
weighted_quantile <- Hmisc::wtd.quantile(
x = dep_var,
weights = weights,
probs = top_share
)
lorenz_ordinate <- sum(dep_var[which(dep_var <= weighted_quantile)] *
weights[which(dep_var <= weighted_quantile)]) /
sum(dep_var * weights)
if_lorenz_ordinate <- -(dep_var / weighted_mean) * lorenz_ordinate +
ifelse(dep_var < weighted_quantile,
dep_var - (1 - top_share) * weighted_quantile,
top_share * weighted_quantile
) / weighted_mean
rif_top_income_share <- (1 - lorenz_ordinate) - if_lorenz_ordinate
rif <- data.frame(rif_top_income_share, weights)
names(rif) <- c("rif_top_income_share", "weights")
return(rif)
}
custom_decomposition <- ob_decompose(
formula = model_rifreg,
data = men8305,
group = year,
reweighting = TRUE,
rifreg_statistic = "custom",
custom_rif_function = custom_top_inc_share,
bootstrap = TRUE,
cores = 4
)
plot(custom_decomposition)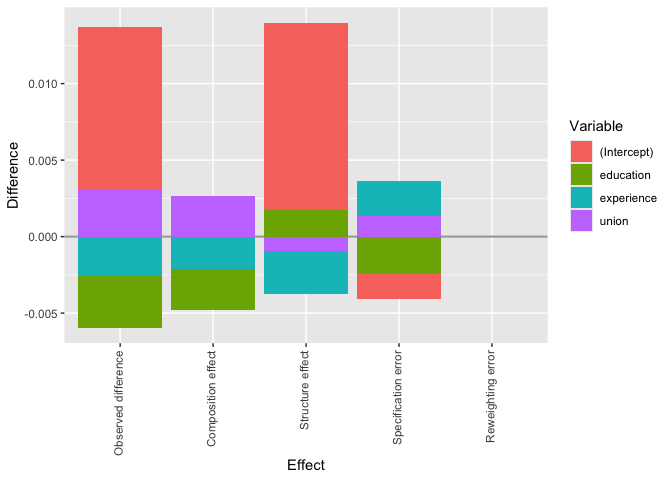 The
plot shows the observed or overall difference of each covariate, which
adds up to the total change in the top ten percent income share from
1983/85 to 2003/05. The composition effect indicates the part
attributable to changes in education, experience and union status, while
the structure effect indicates the part attributable to the returns to
these covariates. The very small reweighting error indicates that the
reweighting worked, while the relatively large specification error
suggests that wage model could be improved.
The
plot shows the observed or overall difference of each covariate, which
adds up to the total change in the top ten percent income share from
1983/85 to 2003/05. The composition effect indicates the part
attributable to changes in education, experience and union status, while
the structure effect indicates the part attributable to the returns to
these covariates. The very small reweighting error indicates that the
reweighting worked, while the relatively large specification error
suggests that wage model could be improved.
Now, we present the use of the other main function:
dfl_decompose(). Fortin, Lemieux, and Firpo (FLF, 2011,
pp. 79-88) decompose the increase in U.S. male wage inequality between
the early 1980s and the early 2000s using the CPS data. In this example,
we perform the same decomposition on subsample of the original data. We
treat the observations from 1983 to 1985 as the reference group, which
is then reweighted. The formula defines on the left-hand side the
outcome variable and specifies on the right-hand side the conditional
probability model.
data("men8305")
flf_model <- log(wage) ~ union * (education + experience) +
education * experience
flf_male_inequality <- dfl_decompose(
formula = flf_model,
data = men8305,
weights = weights,
group = year,
bootstrap = TRUE
)We can summarize the results:
summary(flf_male_inequality)
#> Decomposition of difference between year == '2003-2005' (group 1) and
#> year == '1983-1985' (group 0)
#>
#> Reweighted reference group: year == '1983-1985' (group 0)
#>
#> Composition effect accounts for between-group differences
#> in the distribution of the following covariates:
#>
#> union, education, experience
#>
#> -----------------------------------------------------------------------
#> Decomposition of difference at conditional quantiles:
#>
#> Observed difference:
#> -----------------------------------------------------------------------
#> Quantile Estimate Std. Error [ Pointwise 95%-CI ] [ Uniform 95%-CI ]
#> 0.1 0.062533 0.008564 0.045747 0.07932 0.030074 0.09499
#> 0.2 0.031163 0.009477 0.012588 0.04974 -0.004756 0.06708
#> 0.3 0.007079 0.011295 -0.015058 0.02922 -0.035728 0.04989
#> 0.4 -0.002060 0.010787 -0.023202 0.01908 -0.042942 0.03882
#> 0.5 0.004041 0.007064 -0.009804 0.01788 -0.022730 0.03081
#> 0.6 0.022516 0.007446 0.007923 0.03711 -0.005703 0.05074
#> 0.7 0.044445 0.010408 0.024046 0.06484 0.005000 0.08389
#> 0.8 0.101837 0.009430 0.083354 0.12032 0.066096 0.13758
#> 0.9 0.174073 0.011051 0.152412 0.19573 0.132189 0.21596
#>
#> Composition effect:
#> -----------------------------------------------------------------------
#> Quantile Estimate Std. Error [ Pointwise 95%-CI ] [ Uniform 95%-CI ]
#> 0.1 0.03342 0.005910 0.02184 0.04500 0.01399 0.05285
#> 0.2 0.05429 0.006787 0.04099 0.06759 0.03198 0.07660
#> 0.3 0.07207 0.007469 0.05743 0.08671 0.04752 0.09662
#> 0.4 0.07697 0.006496 0.06424 0.08970 0.05562 0.09832
#> 0.5 0.09504 0.006861 0.08159 0.10849 0.07249 0.11759
#> 0.6 0.08023 0.005876 0.06871 0.09175 0.06092 0.09954
#> 0.7 0.08993 0.005659 0.07884 0.10102 0.07133 0.10853
#> 0.8 0.11981 0.006322 0.10742 0.13220 0.09903 0.14059
#> 0.9 0.12265 0.007312 0.10832 0.13699 0.09862 0.14669
#>
#> Structure effect:
#> -----------------------------------------------------------------------
#> Quantile Estimate Std. Error [ Pointwise 95%-CI ] [ Uniform 95%-CI ]
#> 0.1 0.02911 0.005666 0.01801 0.040220 0.003544 0.054684
#> 0.2 -0.02313 0.008889 -0.04055 -0.005707 -0.063239 0.016982
#> 0.3 -0.06499 0.009468 -0.08355 -0.046435 -0.107718 -0.022266
#> 0.4 -0.07903 0.008638 -0.09596 -0.062100 -0.118007 -0.040052
#> 0.5 -0.09100 0.006902 -0.10453 -0.077471 -0.122146 -0.059852
#> 0.6 -0.05771 0.006619 -0.07069 -0.044740 -0.087582 -0.027844
#> 0.7 -0.04548 0.009524 -0.06415 -0.026815 -0.088462 -0.002502
#> 0.8 -0.01797 0.010034 -0.03764 0.001691 -0.063251 0.027303
#> 0.9 0.05142 0.012701 0.02653 0.076311 -0.005895 0.108731
#>
#> Decomposition of difference for other distributional statistics
#>
#> Observed difference:
#> -----------------------------------------------------------------------
#> Statistic Estimate Std. Error [ 95%-CI ]
#> Mean 0.06205 0.006143 0.05001 0.07410
#> Variance 0.06211 0.004228 0.05383 0.07040
#> Gini* 0.04299 0.002119 0.03884 0.04715
#> Interquantile range p90-p10 0.11154 0.012804 0.08644 0.13663
#> Interquantile range p90-p50 0.17003 0.010772 0.14892 0.19115
#> Interquantile range p50-p10 -0.05849 0.008871 -0.07588 -0.04111
#>
#> Composition effect:
#> -----------------------------------------------------------------------
#> Statistic Estimate Std. Error [ 95%-CI ]
#> Mean 0.080587 0.004439 0.071886 0.089288
#> Variance 0.023498 0.002158 0.019268 0.027729
#> Gini* 0.007114 0.001066 0.005025 0.009203
#> Interquantile range p90-p10 0.089236 0.008736 0.072112 0.106359
#> Interquantile range p90-p50 0.027615 0.007663 0.012595 0.042635
#> Interquantile range p50-p10 0.061621 0.007655 0.046617 0.076624
#>
#> Structure effect:
#> -----------------------------------------------------------------------
#> Statistic Estimate Std. Error [ 95%-CI ]
#> Mean -0.01853 0.005795 -0.029890 -0.007175
#> Variance 0.03861 0.004379 0.030031 0.047195
#> Gini* 0.03588 0.002069 0.031825 0.039937
#> Interquantile range p90-p10 0.02230 0.012858 -0.002898 0.047506
#> Interquantile range p90-p50 0.14242 0.011925 0.119044 0.165790
#> Interquantile range p50-p10 -0.12011 0.008410 -0.136596 -0.103630
#>
#> *Gini of untransformed Y (=exp(log(Y)))
#>
#> -----------------------------------------------------------------------
#> Summary statistics of reweighting factors
#>
#> Number of trimmed observations (not included in statistics): 0 (0%)
#>
#> Psi_X1:
#> -----------------------------------------------------------------------
#> Estimate Std. Error
#> Min. 0.1179 NA
#> 10%-quantile 0.3407 0.02641
#> 25%-quantile 0.6318 0.02304
#> 50%-quantile 0.7669 0.03535
#> 75%-quantile 1.2396 0.04400
#> 90%-quantile 1.7638 0.06849
#> Max. 3.2585 NAUsing plot(), we can illustrate the decomposition across
different quantiles.
plot(flf_male_inequality)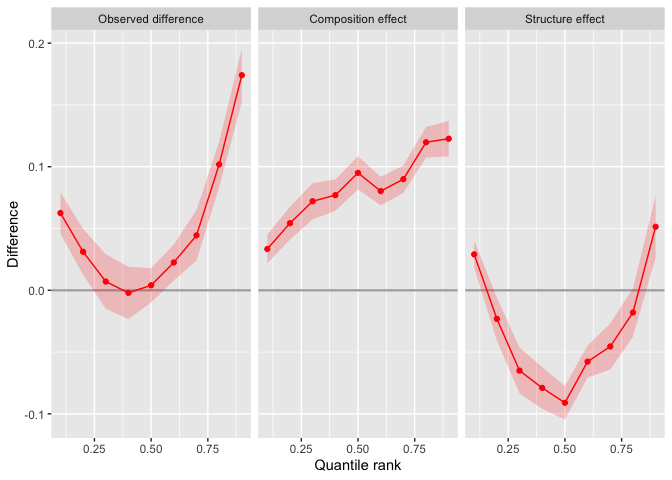
By default estimate_normalized_difference = TRUE and the
normalized differences between the covariate means of the reweighted
reference group and the targeted comparison group are estimated.
head(flf_male_inequality$normalized_difference[[1]])
#> Mean 2003-2005 Mean 1983-1985 (reweighted) SD 2003-2005
#> unionyes 0.15043941 0.14988007 0.3575106
#> educationElementary 0.04725062 0.04721410 0.2121798
#> educationHS dropout 0.08279042 0.08264882 0.2755721
#> educationSome College 0.27481171 0.27534308 0.4464305
#> educationCollege 0.19476140 0.19474447 0.3960268
#> educationPost-graduate 0.09808061 0.09797466 0.2974311
#> SD 1983-1985 (reweighted) Normalized difference
#> unionyes 0.3569626 1.107130e-03
#> educationElementary 0.2121017 1.217075e-04
#> educationHS dropout 0.2753574 3.634824e-04
#> educationSome College 0.4466979 -8.413841e-04
#> educationCollege 0.3960136 3.022945e-05
#> educationPost-graduate 0.2972877 2.519542e-04If we want to further decompose the aggregate decomposition effect
into the contribution of single covariates we can perform a sequential
reweighting decomposition. In the following, we once again decompose the
changes in the US male wage distribution between the early 1980s and the
early 2000s. This time, we want to distinguish the contribution of
changes in the (conditional) unionization from changes in the human
capital endowment. We therefore specify two conditional probability
models in the formula object separated by the | operator.
Beginning from the right, the first model comprises only the human
capital variables (variables \(X_2\) in
the notation from above), while the second also includes a union status
indicator (variable \(X_1\)).
model_sequential <- log(wage) ~ union * (education + experience) +
education * experience |
education * experienceAs before, we use the observations from the early 1980s as reference group.
male_inequality_sequential <- dfl_decompose(
formula = model_sequential,
data = men8305,
weights = weights,
group = year
)The human capital endowment (“Comp. eff. X”) contributed positively to wage growth across the distribution. However, the unionization conditional on education and experience (“Comp. eff. X1|X2”) was negative for all except the two highest income deciles.
plot(male_inequality_sequential)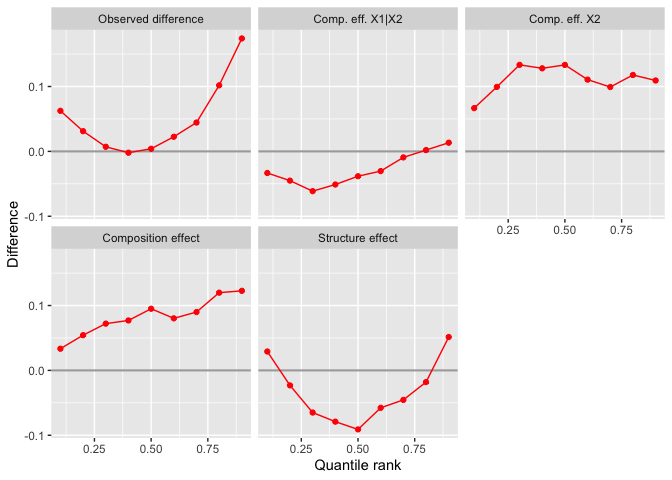 In the example above, we first reweighted the distribution of the early
1980s such that its human capital endowment matches that of the early
2000s. Thus, the counterfactual that identifies the effect of
unionization combines the human capital endowment from the 2000s with
unionization rates from the 1980s.
In the example above, we first reweighted the distribution of the early
1980s such that its human capital endowment matches that of the early
2000s. Thus, the counterfactual that identifies the effect of
unionization combines the human capital endowment from the 2000s with
unionization rates from the 1980s.
Alternatively, we could combine the the human capital endowment from
the 1980s with the unionization rates from the 2000s. In this case, we
have to set the parameter right_to_left = FALSE indicating
that the reweighting begins with the leftmost conditional probability
model in the formula model instead of the rightmost.
male_inequality_sequential_2 <- dfl_decompose(
formula = model_sequential,
data = men8305,
weights = weights,
group = year,
right_to_left = FALSE
)The “direction” of the decomposition set in
right_to_left does not change the aggregate decomposition
results. But the results of the detailed decomposition generally differ
as you can see when comparing the decomposition effects associated with
the (conditional) unionization at the income deciles.
compare_results <- cbind(
male_inequality_sequential$decomposition_quantiles$`Comp. eff. X1|X2`,
male_inequality_sequential_2$decomposition_quantiles$`Comp. eff. X1|X2`
)
colnames(compare_results) <- c("right_to_left = TRUE", "right_to_left = FALSE")
rownames(compare_results) <- male_inequality_sequential$decomposition_quantiles$prob
round(compare_results, 4)
#> right_to_left = TRUE right_to_left = FALSE
#> 0.1 -0.0333 -0.0311
#> 0.2 -0.0451 -0.0318
#> 0.3 -0.0613 -0.0488
#> 0.4 -0.0511 -0.0660
#> 0.5 -0.0383 -0.0605
#> 0.6 -0.0304 -0.0412
#> 0.7 -0.0093 -0.0398
#> 0.8 0.0020 -0.0119
#> 0.9 0.0134 0.0038To validate the functions and provide users with an additional example, we replicate the reweighted RIF regression decomposition estimates in Firpo et al. (2018, p. 30). In their empirical example, Firpo et al. focus on changes in wage inequality in the US between 1988 and 2016. Using a large sample of male log wages in 1988-1990 (268,494 observations) and in 2014-2016 (236,296 observations) based on the Outgoing Rotation Group (ORG) supplement of the Current Population Survey (CPS), the authors attribute changes in several wage inequality measures (i.e, variance, Gini coefficient, and interquantile ranges) between the two time periods to changes in the composition (e.g., union coverage, education, and experience) and changes in the wage structure.
This replication follows the Stata replication code, which the authors published alongside the paper and that is available on one author’s website and here. High wages in the public CPS dataset are top-coded due to privacy concerns. Firpo et al. impute these wages using random draws from a Pareto distribution. Since random numbers are generated differently in R and Stata, even with an equivalent seed, we perform all data preparation in Stata up to the estimation of the decomposition in Stata, i.e., the “oaxaca” commands in Stata. This ensures that changes in the estimation results between our function and the original paper are not due to different input data.
To reproduce the code below, be sure to download the entire dataset and Stata code from the journal’s website, execute the code up to the oaxaca command, save the data, and load it into your R environment.
# Make sure you execute the Stata Code until the "oaxaca" commands
# and save the data in the appropriate folder.
men8816_t4 <- readstata13::read.dta13("data-raw/usmen8816_t4.dta")
# Removing redundant observations - we replicate the reweighting within the function
men8816_t4 <- men8816_t4[men8816_t4$time <= 1, ]The model is specified as in the Stata files. We use sample weights, compute bootstrapped standard errors with 100 iterations, and use the Epanechnikov kernel with a fixed bandwidth of 0.06 in density estimation required to derive the RIF of the interquantile ranges.
set.seed(987421)
ffl_model_with_reweighting <- as.formula(
paste(
"lwage2 ~ covered + nonwhite + nmarr +
ed0 + ed1 + ed3 + ed4 + ed5 + ",
paste(grep(paste0("^ex(", paste(c(1:4, 6:9), collapse = "|"), ")$"),
names(men8816_t4),
value = T
), collapse = " + "), " + ",
paste(grep(paste0("^occd(", paste(c(11:60, 80:91), collapse = "|"), ")$"),
names(men8816_t4),
value = T
), collapse = " + "), " + ",
paste(grep(paste0("^indd(", paste(c(1, 3:14), collapse = "|"), ")$"),
names(men8816_t4),
value = T
), collapse = " + "), " + pub | ",
paste(
"covered + nonwhite +",
paste(grep("^marr",
names(men8816_t4),
value = TRUE
), collapse = " + "), "+",
paste(c("ed0", "ed1", "ed3", "ed4", "ed5"), collapse = " + "), "+",
paste(grep("^ex[1-4]|^ex[6-9]",
names(men8816_t4),
value = TRUE
), collapse = " + "), "+",
paste(grep("^uned",
names(men8816_t4),
value = TRUE
), collapse = " + "), "+",
paste(grep("^unex",
names(men8816_t4),
value = TRUE
), collapse = " + "), "+",
paste(grep("^ex[1-9]ed",
names(men8816_t4),
value = TRUE
), collapse = " + "), "+",
paste(grep("^pub",
names(men8816_t4),
value = TRUE
), collapse = " + "), "+",
paste(grep("^indd(1|1e|[3-9]|10|11|13|14)(?!2)",
names(men8816_t4),
perl = TRUE, value = TRUE
), collapse = " + "), "+",
paste(grep("^occd", names(men8816_t4), value = TRUE), collapse = " + ")
)
)
)
# Interquantile Ratio 90-10
decompose_90_10 <- ob_decompose(
formula = ffl_model_with_reweighting,
data = men8816_t4,
weights = eweight,
group = time,
reference_0 = TRUE,
rifreg_statistic = "interquantile_range",
rifreg_probs = c(0.9, 0.1),
bw = 0.065,
kernel = "epanechnikov",
reweighting = TRUE,
reweighting_method = "fastglm",
trimming = TRUE,
trimming_threshold = 100,
bootstrap = TRUE,
bootstrap_iterations = 100
)
## Variance
set.seed(23904875)
decompose_variance <- ddecompose::ob_decompose(
formula = ffl_model_with_reweighting,
data = men8816_t4,
weights = eweight,
group = time,
reference_0 = TRUE,
rifreg_statistic = "variance",
reweighting = TRUE,
reweighting_method = "fastglm",
trimming = TRUE,
trimming_threshold = 100,
bootstrap = TRUE,
bootstrap_iterations = 100
)
## Gini
# Updating the model
gini_model_raw <- update(ffl_model_with_reweighting, exp(.) ~ .)
gini_model_character <- as.character(gini_model_raw)
gini_model_split <- strsplit(gini_model_character, "~")
ffl_model_with_reweighting_gini <-
as.formula(paste(
gini_model_split[[2]], "~",
gsub("\\(|\\)", "", gini_model_split[[3]][1])
))
set.seed(130234976)
decompose_gini <- ddecompose::ob_decompose(
formula = ffl_model_with_reweighting_gini,
data = men8816_t4,
weights = eweight,
group = time,
rifreg_statistic = "gini",
reweighting = TRUE,
reweighting_method = "fastglm",
trimming = TRUE,
trimming_threshold = 100,
bootstrap = TRUE,
bootstrap_iterations = 100,
cores = 1
)# Presenting the results
variables <- decompose_variance[["variance"]][["decomposition_terms"]][["Variable"]]
ffl_aggregation <- list(
`Union` = "covered",
`Other` = c(
"nonwhite", "nmarr",
grep("ex", variables, value = TRUE)
),
`Education` = grep("ed[0-9]", variables, value = TRUE),
`Occupation` = grep("occd", variables, value = TRUE),
`Industry` = grep("indd", variables, value = TRUE)
)
summary(decompose_90_10, custom_aggregation = ffl_aggregation)
#>
#>
#> Reweighted RIF regression decomposition of difference in interquantile_range
#> between time == '1' (group 1) and time == '0' (group 0).
#> The reference group is '0'.
#>
#> Group 0: time == '0' (0 observations)
#> Group 1: time == '1' (0 observations)
#> Group C: time == '0' (reference group) reweighted
#> to match the characteristics of the other group (0 observations).
#>
#> Pure Composition Effect: (XC - X0) * b0
#> Pure Structure Effect: XC * (bC - b0)
#> Specification Error: (X1 - XC) * bC
#> Reweighting Error: X1 * (b1 - bC)
#>
#> Aggregate decomposition:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Observed difference 0.1256140541 0.004701494 0.116399295 0.1348288131
#> Composition effect 0.0909463043 0.002916652 0.085229771 0.0966628379
#> Structure effect 0.0381813178 0.005043205 0.028296818 0.0480658174
#> Specification error -0.0005160126 0.003691050 -0.007750338 0.0067183127
#> Reweighting error -0.0029975554 0.001399051 -0.005739645 -0.0002554653
#>
#>
#> Observed difference:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.02977302 0.001658415 0.02652259 0.033023458
#> Other -0.02891464 0.010296904 -0.04909620 -0.008733084
#> Education 0.02497305 0.004515913 0.01612202 0.033824073
#> Occupation 0.06949078 0.007782856 0.05423666 0.084744897
#> Industry -0.04775471 0.010027084 -0.06740744 -0.028101989
#> (Other variables) 0.07804656 0.014766600 0.04910456 0.106988565
#>
#>
#> Pure structure effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.012603651 0.001468132 0.009726165 0.01548114
#> Other -0.046989402 0.012405103 -0.071302956 -0.02267585
#> Education 0.058884186 0.006324962 0.046487489 0.07128088
#> Occupation 0.009694848 0.009540193 -0.009003587 0.02839328
#> Industry -0.079592423 0.011883520 -0.102883695 -0.05630115
#> (Other variables) 0.083580458 0.017346762 0.049581429 0.11757949
#>
#>
#> Pure composition effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.016196176 0.0006300468 0.014961307 0.017431045
#> Other 0.019067346 0.0012139205 0.016688106 0.021446587
#> Education 0.008089822 0.0015352464 0.005080794 0.011098850
#> Occupation 0.021190773 0.0013468292 0.018551036 0.023830510
#> Industry 0.024753937 0.0013971957 0.022015483 0.027492390
#> (Other variables) 0.001648250 0.0002679935 0.001122993 0.002173508
#>
#>
#> Specification error:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.0008615798 0.0008615798 -0.0008270856 0.002550245
#> Other -0.0010756721 -0.0010756721 0.0010326065 -0.003183951
#> Education -0.0423315700 -0.0423315700 0.0406367826 -0.125299922
#> Occupation 0.0410382285 0.0410382285 -0.0393952214 0.121471678
#> Industry 0.0082166703 0.0082166703 -0.0078877076 0.024321048
#> (Other variables) -0.0072252492 -0.0072252492 0.0069359791 -0.021386478
#>
#>
#> Reweighting error:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 1.116175e-04 1.116175e-04 -1.071488e-04 0.0003303839
#> Other 8.308368e-05 8.308368e-05 -7.975734e-05 0.0002459247
#> Education 3.306077e-04 3.306077e-04 -3.173715e-04 0.0009785869
#> Occupation -2.433069e-03 -2.433069e-03 2.335659e-03 -0.0072017978
#> Industry -1.132897e-03 -1.132897e-03 1.087540e-03 -0.0033533329
#> (Other variables) 4.310162e-05 4.310162e-05 -4.137601e-05 0.0001275793
#>
#> Summary statistics of reweighting factors
#>
#> Number of trimmed observations (not included in statistics): 0 (0%)
#>
#> Psi_X1
#> Min. 0.01240973
#> 10%-quantile 0.24044303
#> 25%-quantile 0.42188219
#> 50%-quantile 0.73438754
#> 75%-quantile 1.25697488
#> 90%-quantile 2.00825420
#> Max. 22.41294385
summary(decompose_variance, custom_aggregation = ffl_aggregation)
#>
#>
#> Reweighted RIF regression decomposition of difference in variance
#> between time == '1' (group 1) and time == '0' (group 0).
#> The reference group is '0'.
#>
#> Group 0: time == '0' (0 observations)
#> Group 1: time == '1' (0 observations)
#> Group C: time == '0' (reference group) reweighted
#> to match the characteristics of the other group (0 observations).
#>
#> Pure Composition Effect: (XC - X0) * b0
#> Pure Structure Effect: XC * (bC - b0)
#> Specification Error: (X1 - XC) * bC
#> Reweighting Error: X1 * (b1 - bC)
#>
#> Aggregate decomposition:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Observed difference 0.077751372 0.0017482219 0.074324920 0.081177823
#> Composition effect 0.042647761 0.0012056833 0.040284665 0.045010857
#> Structure effect 0.033735963 0.0020922369 0.029635254 0.037836672
#> Specification error 0.002737420 0.0008451794 0.001080899 0.004393941
#> Reweighting error -0.001369773 0.0007649579 -0.002869063 0.000129517
#>
#>
#> Observed difference:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 1.150440e-02 0.000717005 0.01009910 0.012909707
#> Other -4.375222e-06 0.005854285 -0.01147856 0.011469812
#> Education 2.149355e-02 0.002355699 0.01687646 0.026110634
#> Occupation 6.005131e-02 0.003420293 0.05334766 0.066754957
#> Industry -2.046241e-02 0.005448059 -0.03114041 -0.009784408
#> (Other variables) 5.168896e-03 0.007777351 -0.01007443 0.020412223
#>
#>
#> Pure structure effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.003532492 0.0007291712 0.002103343 0.004961642
#> Other -0.009084234 0.0065988197 -0.022017683 0.003849215
#> Education 0.024947096 0.0032184078 0.018639133 0.031255059
#> Occupation 0.025238396 0.0040670528 0.017267119 0.033209673
#> Industry -0.034523565 0.0063051274 -0.046881387 -0.022165742
#> (Other variables) 0.023625777 0.0085958611 0.006778199 0.040473356
#>
#>
#> Pure composition effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.007084019 0.0002531646 0.006587826 0.007580213
#> Other 0.010060413 0.0005488449 0.008984697 0.011136129
#> Education 0.006374034 0.0005910651 0.005215567 0.007532500
#> Occupation 0.007480422 0.0005493005 0.006403812 0.008557031
#> Industry 0.010320617 0.0005818809 0.009180152 0.011461083
#> (Other variables) 0.001328256 0.0001487646 0.001036683 0.001619830
#>
#>
#> Specification error:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.0008415311 0.0008415311 -0.0008078395 0.002490902
#> Other -0.0008730819 -0.0008730819 0.0008381272 -0.002584291
#> Education -0.0100362859 -0.0100362859 0.0096344730 -0.029707045
#> Occupation 0.0284495154 0.0284495154 -0.0273105101 0.084209541
#> Industry 0.0041833718 0.0041833718 -0.0040158863 0.012382630
#> (Other variables) -0.0198276304 -0.0198276304 0.0190338111 -0.058689072
#>
#>
#> Reweighting error:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 4.636017e-05 4.636017e-05 -4.450409e-05 0.0001372244
#> Other -1.074720e-04 -1.074720e-04 1.031693e-04 -0.0003181133
#> Education 2.087049e-04 2.087049e-04 -2.003492e-04 0.0006177590
#> Occupation -1.117026e-03 -1.117026e-03 1.072305e-03 -0.0033063578
#> Industry -4.428319e-04 -4.428319e-04 4.251026e-04 -0.0013107664
#> (Other variables) 4.249232e-05 4.249232e-05 -4.079110e-05 0.0001257757
#>
#> Summary statistics of reweighting factors
#>
#> Number of trimmed observations (not included in statistics): 0 (0%)
#>
#> Psi_X1
#> Min. 0.01240973
#> 10%-quantile 0.24044303
#> 25%-quantile 0.42188219
#> 50%-quantile 0.73438754
#> 75%-quantile 1.25697488
#> 90%-quantile 2.00825420
#> Max. 22.41294385
summary(decompose_gini, custom_aggregation = ffl_aggregation)
#>
#>
#> Reweighted RIF regression decomposition of difference in gini
#> between time == '1' (group 1) and time == '0' (group 0).
#> The reference group is '0'.
#>
#> Group 0: time == '0' (0 observations)
#> Group 1: time == '1' (0 observations)
#> Group C: time == '0' (reference group) reweighted
#> to match the characteristics of the other group (0 observations).
#>
#> Pure Composition Effect: (XC - X0) * b0
#> Pure Structure Effect: XC * (bC - b0)
#> Specification Error: (X1 - XC) * bC
#> Reweighting Error: X1 * (b1 - bC)
#>
#> Aggregate decomposition:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Observed difference 0.0659908665 0.0015135722 0.063024320 6.895741e-02
#> Composition effect 0.0199765902 0.0009934684 0.018029428 2.192375e-02
#> Structure effect 0.0451291062 0.0017981373 0.041604822 4.865339e-02
#> Specification error 0.0014785630 0.0005137232 0.000471684 2.485442e-03
#> Reweighting error -0.0005933929 0.0003444451 -0.001268493 8.170719e-05
#>
#>
#> Observed difference:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.009960677 0.0005996969 0.0087852925 0.011136061
#> Other 0.003106513 0.0054771363 -0.0076284769 0.013841503
#> Education 0.004133062 0.0017218577 0.0007582833 0.007507842
#> Occupation 0.023964506 0.0023517073 0.0193552446 0.028573768
#> Industry -0.005860137 0.0036927292 -0.0130977536 0.001377479
#> (Other variables) 0.030686245 0.0062595714 0.0184177107 0.042954780
#>
#>
#> Pure structure effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.0022173824 0.0006105551 0.001020716 0.003414048
#> Other -0.0007297231 0.0061460445 -0.012775749 0.011316303
#> Education 0.0126849132 0.0018750239 0.009009934 0.016359893
#> Occupation 0.0130636777 0.0026466873 0.007876266 0.018251090
#> Industry -0.0121351390 0.0041780504 -0.020323967 -0.003946311
#> (Other variables) 0.0300279951 0.0069379047 0.016429952 0.043626039
#>
#>
#> Pure composition effect:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.0063399542 0.0002157312 0.0059171289 0.006762779
#> Other 0.0047845108 0.0005063463 0.0037920903 0.005776931
#> Education 0.0019375518 0.0004602056 0.0010355653 0.002839538
#> Occupation 0.0013849785 0.0004112917 0.0005788616 0.002191095
#> Industry 0.0047148753 0.0004569234 0.0038193219 0.005610429
#> (Other variables) 0.0008147197 0.0001167025 0.0005859871 0.001043452
#>
#>
#> Specification error:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 0.0013650891 0.0013650891 -0.0013104364 0.0040406147
#> Other -0.0009370702 -0.0009370702 0.0008995537 -0.0027736942
#> Education -0.0105565134 -0.0105565134 0.0101338727 -0.0312468996
#> Occupation 0.0099999030 0.0099999030 -0.0095995468 0.0295993528
#> Industry 0.0017927361 0.0017927361 -0.0017209621 0.0053064343
#> (Other variables) -0.0001855816 -0.0001855816 0.0001781517 -0.0005493149
#>
#>
#> Reweighting error:
#>
#> Estimate Std. Error [ 95%-CI ]
#> Union 3.825111e-05 3.825111e-05 -3.671969e-05 1.132219e-04
#> Other -1.120429e-05 -1.120429e-05 1.075572e-05 -3.316430e-05
#> Education 6.711095e-05 6.711095e-05 -6.442409e-05 1.986460e-04
#> Occupation -4.840530e-04 -4.840530e-04 4.646734e-04 -1.432779e-03
#> Industry -2.326096e-04 -2.326096e-04 2.232969e-04 -6.885162e-04
#> (Other variables) 2.911195e-05 2.911195e-05 -2.794643e-05 8.617033e-05
#>
#> Summary statistics of reweighting factors
#>
#> Number of trimmed observations (not included in statistics): 0 (0%)
#>
#> Psi_X1
#> Min. 0.01240973
#> 10%-quantile 0.24044303
#> 25%-quantile 0.42188219
#> 50%-quantile 0.73438754
#> 75%-quantile 1.25697488
#> 90%-quantile 2.00825420
#> Max. 22.41294385The results presented here are similar to those in Table 4 of Firpo et al. (2018, p. 30). However, some of the coefficients calculated by us differ slightly from the results in the paper. There are several reasons for these differences.
Reweighting: An important difference is the reweighting factors.
Firpo et al. use the entire dataset to compute the reweighting factors.
However, for the decomposition they remove very high wages from the
dataset. In ob_decompose(), the same (trimmed) dataset is
used for reweighting and the decomposition estimation.
Different decomposition formula: In the paper, the formula presented for the pure structure and reweighting errors is identical to the formula presented in the background section above. For instance, the pure wage structure effect is computed as \(\overline X_{1,1}\widehat \beta_{1,1} - \widehat \beta_{C,1}\) for \(k=1\). However, the Stata code calculates a slightly different formula: \(\overline X_{C,1}(\widehat \beta_{C,1} - \widehat \beta_{1,1})\). Thus, in the Stata replication code, the results are multiplied by -1 so that the composition and and structure effects add up to the observed difference. When we calculate the results in Stata, using the formula presented in the paper, our function produces very similar results to the Stata output, with a deviations of less than 0.1 percent in most cases.
Density estimation: For the interquantile ranges the differences
are generally larger. We attribute this to the different density
estimations in Stata and R (even when using the same kernel function and
bandwidth). Specifically, kdensity in Stata and
stats::density() in R set the grids that define the locus
of the density estimates differently. These differences result in
different RIF values and thus different regression
coefficients.
The bootstrapped standard errors are relatively similar to those reported in the paper. With only 100 bootstrap replications and different seeds, some variance in the terms is not surprising. In addition, we also included the reweighting procedure in the bootstrap estimation, while Fortin et al. only include the RIF regression estimation.
In summary, our ob_decompose() function produces very
similar results to those calculated in Stata and presented in the
original paper. Using an identical formula, the deviations are mostly
below 0.1 percent. However, some values based on RIF estimations of
quantiles have slightly higher differences. We have also replicated the
results of Table 1-3 in Firpo et al. (2018, pp. 21-29), where the
differences are generally even smaller. The replication files are
available upon request. This validation example illustrates that the
ddecompose package works as intended in computing
reweighted RIF regression decompositions and reliably produces the
expected results.
David Gallusser & Samuel Meier
Barsky, Robert, John Bound, Kerwin Kofi Charles, and Joseph P. Lupton. 2002. “Accounting for the Black-White Wealth Gap: A Nonparametric Approach.” Journal of the American Statistical Association 97: 663–73.
Cowell, Frank A., and Emmanuel Flachaire. 2007. “Income distribution and inequality measurement: The problem of extreme values.” Journal of Econometrics 141: 1044–1072.
Chen, Mingli, Victor Chernozhukov, Iván Fernández-Val, and Blaise Melly. 2017. “Counterfactual: An R Package for Counterfactual Analysis.” The R Journal 9(1): 370-384.
DiNardo, John, Nicole M. Fortin, and Thomas Lemieux. 1996. “Labor Market Institutions and the Distribution of Wages, 1973-1992: A Semiparametric Approach.” Econometrica 64(5): 1001-1044.
Essama-Nssah, Boniface, and Peter J. Lambert. 2012. “Influence Functions for Policy Impact Analysis.” In John A. Bishop and Rafael Salas, eds., Inequality, Mobility and Segregation: Essays in Honor of Jacques Silber. Bigley, UK: Emerald Group Publishing Limited.
Firpo, Sergio. 2007. “Efficient Semiparametric Estimation of Quantile Treatment Effects.” Econometrica 75(1): 259-276.
Firpo, Sergio, Nicole M. Fortin, and Thomas Lemieux. 2007a. “Unconditional Quantile Regressions.” Technical Working Paper 339, National Bureau of Economic Research. Cambridge, MA.
Firpo, Sergio, Nicole M. Fortin, and Thomas Lemieux. 2009a. “Unconditional Quantile Regressions.” Econometrica 77(3): 953-973.
Firpo, Sergio, Nicole M. Fortin, and Thomas Lemieux. 2009b. “Supplement to ‘Unconditional Quantile Regressions’.” Econometrica Supplemental Material, 77.
Firpo, Sergio, Nicole M. Fortin, and Thomas Lemieux. 2018. “Decomposing Wage Distributions Using Recentered Influence Function Regressions.” Econometrics 6(2):28.
Fortin, Nicole M., Thomas Lemieux, and Sergio Firpo. 2011. “Decomposition Methods in Economics.” National Bureau of Economic Research - Working Paper Series, 16045.
Firpo, Sergio, and Pinto, Christine. 2016. “Identification and Estimation of Distributional Impacts of Interventions Using Changes in Inequality Measures.” Journal of Applied Econometrics, 31(3), 457–486.
Gardeazabal, Javier, and Arantza Ugidos. 2004. “More on identification in detailed wage decompositions.” Review of Economics and Statistics, 86(4): 1034-1036.
Hlavac, Marek. 2022. “oaxaca: Blinder-Oaxaca Decomposition in R”. R package version 0.1.5. https://CRAN.R-project.org/package=oaxaca.
Huber, Martin, Michael Lechner, and Conny Wunsch. 2013. “The performance of estimators based on the propensity score.” Journal of Econometrics 175(1): 1-21.
Jann, Ben. 2005. “Standard errors for the Blinder-Oaxaca decomposition.” 3rd German Stata Users’ Group Meeting 2005. Available from https://boris.unibe.ch/69506/1/oaxaca_se_handout.pdf.
Jann, Ben. 2008. “The Blinder–Oaxaca Decomposition for Linear Regression Models”. Stata Journal 8: 435–479.
Rios-Avila, Fernando. 2020. “Recentered influence functions (RIFs) in Stata: RIF regression and RIF decomposition.” The Stata Journal 20(1): 51-94.
Rothe, Christoph. 2012. “Decomposing the Composition Effect. The Role of Covariates in Determining Between-Group Differences in Economic Outcomes.” IZA Discussion Paper No. 6397.
Rothe, Christoph. 2015. “Decomposing the Composition Effect. The Role of Covariates in Determining Between-Group Differences in Economic Outcomes.” Journal of Business & Economic Statistics 33(3): 323-337.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.