The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
An implementation of Hastie and Tibshirani’s Discriminant Adaptive Nearest Neighbor Classification in R.
In k nearest neighbors, the shape of the neighborhood is usually circular. Discriminant Adaptive Nearest Neighbor (dann) is a variation of k nearest neighbors where the shape of the neighborhood is data driven. The neighborhood is elongated along class boundaries and shrunk in the orthogonal direction. See Discriminate Adaptive Nearest Neighbor Classification by Hastie and Tibshirani.
Models:
This package implements DANN and sub-DANN in section 4.1 of the publication and is based on Christopher Jenness’s python implementation.
Arguments:
In this example, simulated data is made. The overall trend is a circle inside a square.
knitr::opts_chunk$set(echo = TRUE, fig.width = 10, fig.height = 10)
library(dann)
library(dplyr, warn.conflicts = FALSE)
library(ggplot2)
library(mlbench)
set.seed(1)
# Create training data
train <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(train) <- c("X1", "X2", "Y")
train <- train %>%
mutate(Y = as.numeric(Y))
ggplot(train, aes(x = X1, y = X2, colour = as.factor(Y))) +
geom_point() +
labs(title = "Train Data", colour = "Y")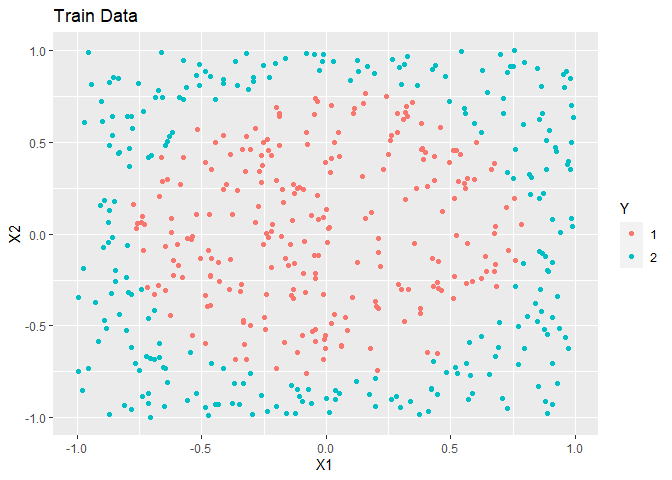
To train a model, call dann.
model <- dann(formula = Y ~ X1 + X2, data = train, k = 5, neighborhood_size = 50, epsilon = 1)To get class predictions, call predict with type equal to “class”
# Create test data
test <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(test) <- c("X1", "X2", "Y")
test <- test %>%
mutate(Y = as.numeric(Y))
yhat <- predict(object = model, new_data = test, type = "class")
yhat
#> # A tibble: 500 × 1
#> .pred_class
#> <fct>
#> 1 2
#> 2 1
#> 3 1
#> 4 2
#> 5 2
#> 6 2
#> 7 1
#> 8 1
#> 9 2
#> 10 1
#> # ℹ 490 more rowsTo get probabilities, call predict with type equal to “prob”
yhat <- predict(object = model, new_data = test, type = "prob")
yhat
#> # A tibble: 500 × 2
#> .pred_1 .pred_2
#> <dbl> <dbl>
#> 1 0 1
#> 2 0.6 0.4
#> 3 1 0
#> 4 0 1
#> 5 0 1
#> 6 0 1
#> 7 1 0
#> 8 1 0
#> 9 0 1
#> 10 1 0
#> # ℹ 490 more rowsIn general, dann will struggle as unrelated variables are intermingled with informative variables. To deal with this, sub_dann projects the data onto a unique subspace and then calls dann on the subspace.
In the below example there are 2 related variables and 5 that are unrelated. Will sub_dann to better than dann?
######################
# Circle data with unrelated variables
######################
set.seed(1)
train <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(train)[1:3] <- c("X1", "X2", "Y")
# Add 5 unrelated variables
train <- train %>%
mutate(
U1 = runif(500, -1, 1),
U2 = runif(500, -1, 1),
U3 = runif(500, -1, 1),
U4 = runif(500, -1, 1),
U5 = runif(500, -1, 1)
)
test <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(test)[1:3] <- c("X1", "X2", "Y")
# Add 5 unrelated variables
test <- test %>%
mutate(
U1 = runif(500, -1, 1),
U2 = runif(500, -1, 1),
U3 = runif(500, -1, 1),
U4 = runif(500, -1, 1),
U5 = runif(500, -1, 1)
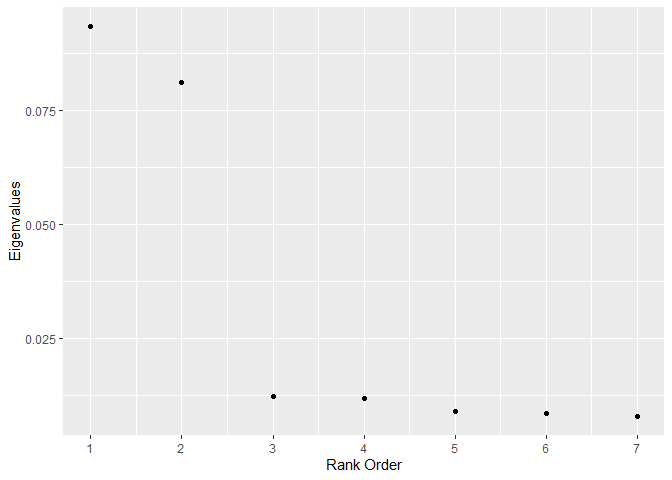
)For sub_dann, the dimension of the subspace should be chosen based on the number of large eigenvalues. The graph suggests 2 (the correct answer).
knitr::opts_chunk$set(echo = TRUE, fig.width = 10, fig.height = 10)
graph_eigenvalues(
formula = Y ~ X1 + X2 + U1 + U2 + U3 + U4 + U5,
data = train,
neighborhood_size = 50,
weighted = FALSE,
sphere = "mcd"
)
dann_model <- dann(
formula = Y ~ X1 + X2 + U1 + U2 + U3 + U4 + U5,
data = train,
k = 3,
neighborhood_size = 50,
epsilon = 1
)
# numDim based on large eigenvalues
# weighted, sphere, and neighborhood_size kept consistent between sub_dann and graph_eigenvalues
sub_dann_model <- sub_dann(
formula = Y ~ X1 + X2 + U1 + U2 + U3 + U4 + U5,
data = train,
k = 3,
neighborhood_size = 50,
epsilon = 1,
weighted = FALSE,
sphere = "mcd",
numDim = 2
)As expected, the dann model using all features does not fit the data well.
library(yardstick)
dann_yhat <- predict(object = dann_model, new_data = test, type = "prob")
dann_yhat <- test %>%
select(Y) %>%
bind_cols(dann_yhat)
roc_auc(data = dann_yhat, truth = Y, event_level = "first", .pred_1)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.725sub_dann provides a major improvement.
sub_dann_yhat <- predict(object = sub_dann_model, new_data = test, type = "prob")
sub_dann_yhat <- test %>%
select(Y) %>%
bind_cols(sub_dann_yhat)
roc_auc(data = sub_dann_yhat, truth = Y, event_level = "first", .pred_1)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.935These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.