The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The goal of cvwrapr is to make cross-validation (CV)
easy. The main function in the package is kfoldcv. It
performs K-fold CV for a hyperparameter, returning the CV error for a
path of hyperparameter values along with other useful information. The
computeError function allows the user to compute the CV
error for a range of loss functions from a matrix of out-of-fold
predictions. See the package vignettes for more examples.
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("kjytay/cvwrapr")This is a basic example showing how to perform cross-validation for
the lambda parameter in the lasso (Tibshirani 1996).
# simulate data
set.seed(1)
nobs <- 100; nvars <- 10
x <- matrix(rnorm(nobs * nvars), nrow = nobs)
y <- rowSums(x[, 1:2]) + rnorm(nobs)
library(cvwrapr)
library(glmnet)
set.seed(1)
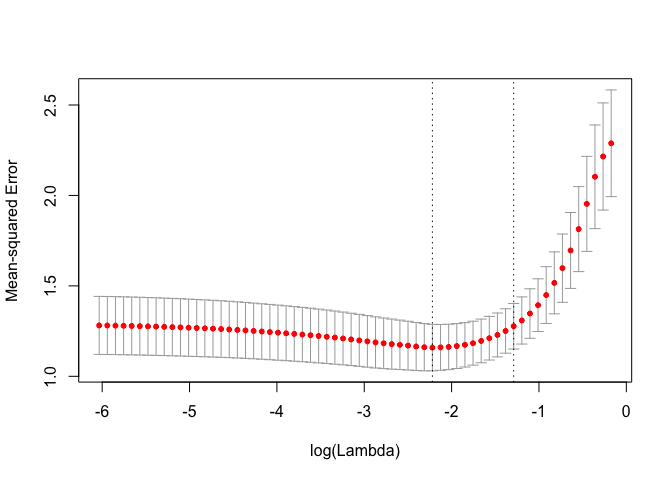
cv_fit <- kfoldcv(x, y, train_fun = glmnet, predict_fun = predict)The returned output contains information on the CV procedure and can be plotted.
names(cv_fit)
#> [1] "lambda" "cvm" "cvsd" "cvup" "cvlo"
#> [6] "lambda.min" "lambda.1se" "index" "name" "overallfit"
plot(cv_fit)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.