The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
An R package for
composable maximum likelihood estimation. Solvers are
first-class functions that combine via sequential chaining, parallel
racing, and random restarts.
Use compositional.mle when:
Stick with optim() when:
library(compositional.mle)
# A tricky bimodal likelihood
set.seed(42)
bimodal_loglike <- function(theta) {
# Two peaks: one at theta=2, one at theta=8
log(0.3 * dnorm(theta, 2, 0.5) + 0.7 * dnorm(theta, 8, 0.5))
}
problem <- mle_problem(
loglike = bimodal_loglike,
constraint = mle_constraint(support = function(theta) TRUE)
)
# Single gradient ascent gets trapped at local optimum
result_local <- gradient_ascent()(problem, theta0 = 0)
# Simulated annealing + gradient ascent finds global optimum
strategy <- sim_anneal(temp_init = 5, max_iter = 200) %>>% gradient_ascent()
result_global <- strategy(problem, theta0 = 0)
cat("Local search found:", round(result_local$theta.hat, 2),
"(log-lik:", round(result_local$loglike, 2), ")\n")
#> Local search found: 2 (log-lik: -1.43 )
cat("Global strategy found:", round(result_global$theta.hat, 2),
"(log-lik:", round(result_global$loglike, 2), ")\n")
#> Global strategy found: 2 (log-lik: -1.43 )# From CRAN (when available)
install.packages("compositional.mle")
# Development version
devtools::install_github("queelius/compositional.mle")Following SICP principles, the package provides: 1. Primitive
solvers - gradient_ascent(),
newton_raphson(), bfgs(),
sim_anneal(), etc. 2. Composition
operators - %>>% (sequential),
%|% (race), with_restarts() 3. Closure
property - Combining solvers yields a solver
# Generate sample data
set.seed(42)
x <- rnorm(100, mean = 5, sd = 2)
# Define the problem (separate from solver strategy)
problem <- mle_problem(
loglike = function(theta) {
if (theta[2] <= 0) return(-Inf)
sum(dnorm(x, theta[1], theta[2], log = TRUE))
},
score = function(theta) {
mu <- theta[1]; sigma <- theta[2]; n <- length(x)
c(sum(x - mu) / sigma^2,
-n / sigma + sum((x - mu)^2) / sigma^3)
},
constraint = mle_constraint(
support = function(theta) theta[2] > 0,
project = function(theta) c(theta[1], max(theta[2], 1e-8))
)
)
# Simple solve
result <- gradient_ascent()(problem, theta0 = c(0, 1))
result$theta.hat
#> [1] 5.065030 2.072274%>>%)Chain solvers for coarse-to-fine optimization:
# Grid search -> gradient ascent -> Newton-Raphson
strategy <- grid_search(lower = c(-10, 0.5), upper = c(10, 5), n = 5) %>>%
gradient_ascent(max_iter = 50) %>>%
newton_raphson(max_iter = 20)
result <- strategy(problem, theta0 = c(0, 1))
result$theta.hat
#> Var1 Var2
#> 5.065030 2.072274%|%)Race multiple methods, keep the best:
# Try multiple approaches, pick winner by log-likelihood
strategy <- gradient_ascent() %|% bfgs() %|% nelder_mead()
result <- strategy(problem, theta0 = c(0, 1))
c(result$theta.hat, loglike = result$loglike)
#> loglike
#> 5.065030 2.072274 -214.758518Escape local optima with multiple starting points:
strategy <- with_restarts(
gradient_ascent(),
n = 10,
sampler = uniform_sampler(c(-10, 0.5), c(10, 5))
)
result <- strategy(problem, theta0 = c(0, 1))
result$theta.hat
#> [1] 5.065030 2.072274Track and visualize the optimization path:
# Enable tracing
trace_cfg <- mle_trace(values = TRUE, gradients = TRUE, path = TRUE)
result <- gradient_ascent(max_iter = 50)(problem, c(0, 1), trace = trace_cfg)
# Plot convergence
plot(result, which = c("loglike", "gradient"))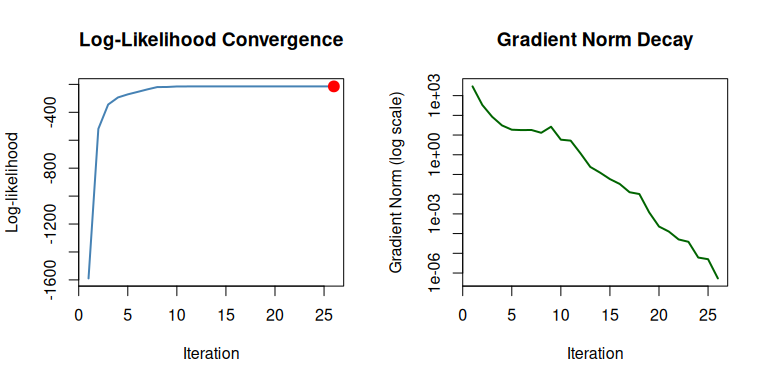
Extract trace as data frame for custom analysis:
path_df <- optimization_path(result)
head(path_df)
#> iteration loglike grad_norm theta_1 theta_2
#> 1 1 -1589.3361 2938.86063 0.0000000 1.000000
#> 2 2 -518.7066 334.47435 0.1723467 1.985036
#> 3 3 -344.5384 84.57555 0.5435804 2.913576
#> 4 4 -293.8366 30.83372 1.1733478 3.690360
#> 5 5 -271.7336 18.65296 2.1001220 4.065978
#> 6 6 -254.0618 17.83778 3.0615865 3.791049| Factory | Method | Best For |
|---|---|---|
gradient_ascent() |
Steepest ascent with line search | General purpose, smooth likelihoods |
newton_raphson() |
Second-order Newton | Fast convergence near optimum |
bfgs() |
Quasi-Newton BFGS | Good balance of speed/robustness |
lbfgsb() |
L-BFGS-B with box constraints | High-dimensional, bounded parameters |
nelder_mead() |
Simplex (derivative-free) | Non-smooth or noisy likelihoods |
sim_anneal() |
Simulated annealing | Global optimization, multi-modal |
coordinate_ascent() |
One parameter at a time | Different parameter scales |
grid_search() |
Exhaustive grid | Finding starting points |
random_search() |
Random sampling | High-dimensional exploration |
# Stochastic gradient (mini-batching for large data)
loglike_sgd <- with_subsampling(loglike, data = x, subsample_size = 32)
# Regularization
loglike_l2 <- with_penalty(loglike, penalty_l2(), lambda = 0.1)
loglike_l1 <- with_penalty(loglike, penalty_l1(), lambda = 0.1)MIT
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.