The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
cfr is an R package to estimate disease severity and under-reporting in real-time, accounting for delays in epidemic time-series.
cfr provides simple, fast methods to calculate the overall or static case fatality risk (CFR) of an outbreak up to a given time point, as well as how the CFR changes over the course of the outbreak. cfr can help estimate disease under-reporting in real-time, accounting for delays reporting the outcomes of cases.
cfr implements methods outlined in Nishiura et al. (2009). There are plans to add estimates based on other methods.
cfr is developed at the Centre for the Mathematical Modelling of Infectious Diseases at the London School of Hygiene and Tropical Medicine as part of the Epiverse-TRACE initiative.
cfr can be installed from CRAN using
install.packages("cfr")The current development version of cfr can be installed from
GitHub using the pak
package.
if(!require("pak")) install.packages("pak")
pak::pak("epiverse-trace/cfr")This example shows how to use cfr to estimate the overall case fatality risks from the 1976 Ebola outbreak (Camacho et al. 2014), while correcting for delays using a Gamma-distributed onset to death duration taken from Barry et al. (2018), with a shape \(k\) of 2.40 and a scale \(\theta\) of 3.33.
# Load package
library(cfr)
# Load the Ebola 1976 data provided with the package
data(ebola1976)
# Focus on the first 20 days the outbreak
ebola1976_first_30 <- ebola1976[1:30, ]
# Calculate the static CFR without correcting for delays
cfr_static(data = ebola1976_first_30)
#> severity_estimate severity_low severity_high
#> 1 0.4740741 0.3875497 0.5617606
# Calculate the static CFR while correcting for delays
cfr_static(
data = ebola1976_first_30,
delay_density = function(x) dgamma(x, shape = 2.40, scale = 3.33)
)
#> severity_estimate severity_low severity_high
#> 1 0.9422 0.8701 0.9819In this example we show how the estimate of overall severity can
change as more data on cases and deaths over time becomes available,
using the function cfr_rolling(). Because there is a delay
from onset-to-death, a simple “naive” calculation that just divides
deaths-to-date by cases-to-date will underestimate severity. The
cfr_rolling() function uses the
estimate_severity() adjustment internally to account for
delays, and instead compares deaths-to-date with
cases-with-known-outcome-to-date. The adjusted estimate converges to the
naive estimate as the outbreak declines and a larger proportion of cases
have known outcomes.
# Calculate the CFR without correcting for delays on each day of the outbreak
rolling_cfr_naive <- cfr_rolling(
data = ebola1976
)
# see the first few rows
head(rolling_cfr_naive)
#> date severity_estimate severity_low severity_high
#> 1 1976-08-25 0 0 0.975
#> 2 1976-08-26 0 0 0.975
#> 3 1976-08-27 0 0 0.975
#> 4 1976-08-28 0 0 0.975
#> 5 1976-08-29 0 0 0.975
#> 6 1976-08-30 0 0 0.975
# Calculate the rolling daily CFR while correcting for delays
rolling_cfr_corrected <- cfr_rolling(
data = ebola1976,
delay_density = function(x) dgamma(x, shape = 2.40, scale = 3.33)
)
head(rolling_cfr_corrected)
#> date severity_estimate severity_low severity_high
#> 1 1976-08-25 NA NA NA
#> 2 1976-08-26 1e-04 1e-04 0.9999
#> 3 1976-08-27 1e-04 1e-04 0.9999
#> 4 1976-08-28 1e-04 1e-04 0.9999
#> 5 1976-08-29 1e-04 1e-04 0.9990
#> 6 1976-08-30 1e-04 1e-04 0.9942We plot the rolling CFR to visualise how severity changes over time, using the ggplot2 package. The plotting code is hidden here.
# combine the data for plotting
rolling_cfr_naive$method <- "naive"
rolling_cfr_corrected$method <- "corrected"
data_cfr <- rbind(
rolling_cfr_naive,
rolling_cfr_corrected
)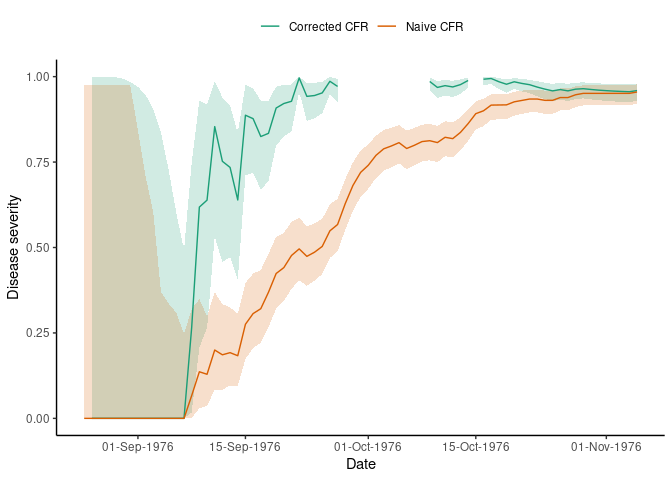
Disease severity of ebola in the 1976 outbreak estimated on each day of the epidemic. The rolling CFR value converges to the static value towards the end of the outbreak. Both corrected and uncorrected estimates are shown.
More details on how to use cfr can be found in the online documentation as package vignettes, under “Articles”.
To report a bug please open an issue.
Contributions to cfr are welcomed. Please follow the package contributing guide.
Please note that the cfr project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
cfr functionality overlaps with that of some other packages, including
cfr is in future expected to benefit from the functionality
of the forthcoming epiparameter
package, which is also developed by Epiverse-TRACE.
epiparameter aims to provide a library of epidemiological
parameters to parameterise delay density functions, as well as the
convenient <epiparameter> class to store, access, and
pass these parameters for delay correction.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.