
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Causal Distillation Trees (CDT) is a novel machine learning method for estimating interpretable subgroups in causal inference. CDT allows researchers to fit any machine learning model of their choice to estimate the individual-level treatment effect, and then leverages a simple, second-stage tree-based model to “distill” the estimated treatment effect into meaningful subgroups. As a result, CDT inherits the improvements in predictive performance from black-box machine learning models while preserving the interpretability of a simple decision tree.

Briefly, CDT is a two-stage learner that first fits a teacher model (e.g., a black-box metalearner) to estimate individual-level treatment effects and secondly fits a student model (e.g., a decision tree) to predict the estimated individual-level treatment effects, in effect distilling the estimated individual-level treatment effects and producing interpretable subgroups. This two-stage learner is learned using the training data. Finally, using the estimated subgroups, the subgroup average treatment effects are honestly estimated with a held-out estimation set.
For more details, check out Huang, M., Tang, T. M., Kenney, A. M. “Distilling heterogeneous treatment effects: Stable subgroup estimation in causal inference.” (2025).
You can install the causalDT R package via:
# install.packages("devtools")
devtools::install_github("tiffanymtang/causalDT", subdir = "causalDT")To illustrate an example usage of causalDT, we will use
the AIDS Clinical Trials Group Study 175 (ACTG 175), a randomized
controlled trial to determine the effectiveness of monotherapy compared
to combination therapy on HIV-1-infected patients. This data can be
found in the speff2trial R package.
# install.packages("speff2trial")
library(speff2trial)
#> Loading required package: leaps
#> Loading required package: survival
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
data <- speff2trial::ACTG175 |>
dplyr::filter(arms %in% c(0, 2))
# pre-treatment covariates data
X <- data |>
dplyr::select(
age, wtkg, hemo, homo, drugs, karnof, race,
gender, symptom, preanti, strat, cd80
) |>
as.matrix()
# treatment indicator variable
Z <- data |>
dplyr::pull(treat)
# response variable
Y <- data |>
dplyr::pull(cens)Given the pre-treatment covariates data \(X\), the treatment variable \(Z\), and the response variable \(Y\), we can run CDT as follows:
library(causalDT)
set.seed(331)
causal_forest_cdt <- causalDT(
X = X, Y = Y, Z = Z,
teacher_model = "causal_forest"
)
plot_cdt(causal_forest_cdt)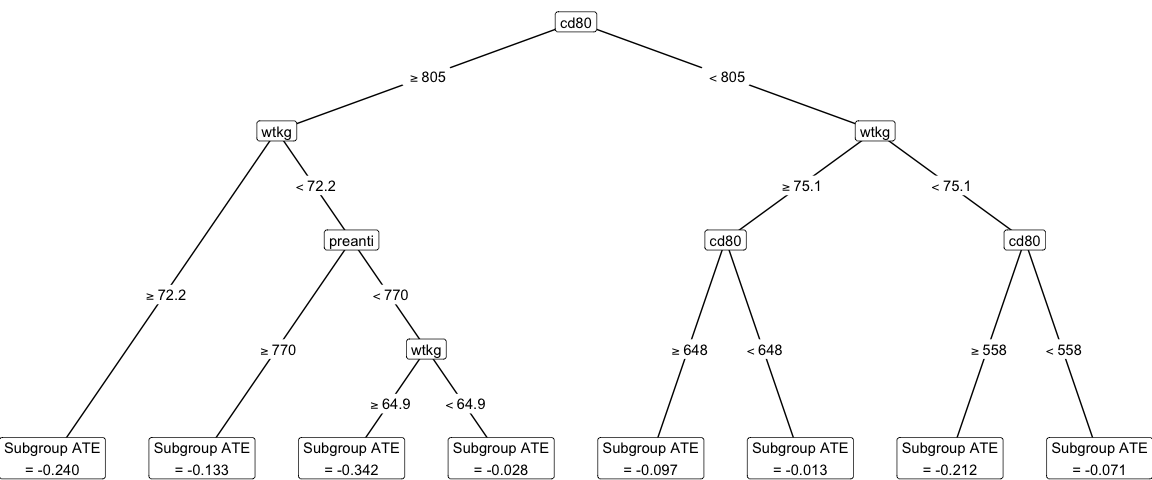
Note that when using CDT, a teacher model must be chosen (the default is a causal forest). To help researchers select an appropriate teacher model, the Jaccard subgroup stability index (SSI) was introduced in Huang et al. (2025). Generally, a higher Jaccard SSI indicates a better teacher model. This teacher model selection procedure can be run as follows:
## uncomment to install rlearner, which is needed to run rboost
# remotes::install_github("xnie/rlearner")
# selecting between causal forest versus rboost
rboost_cdt <- causalDT(
X = as.matrix(X), Y = Y, Z = Z,
teacher_model = rlearner_teacher(rlearner::rboost)
)
plot_jaccard(`Causal Forest` = causal_forest_cdt, `Rboost` = rboost_cdt)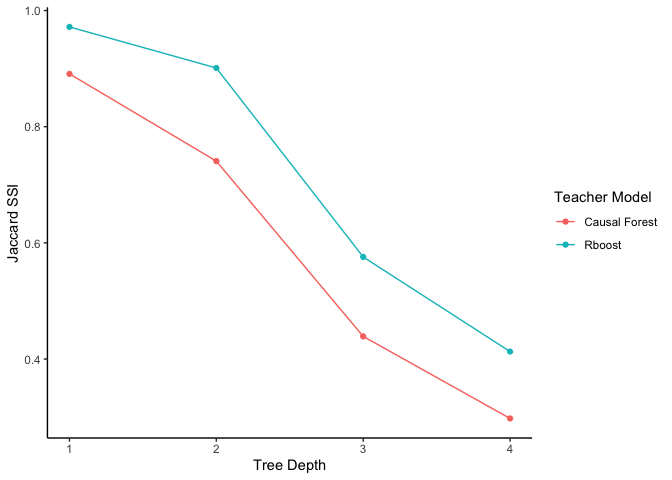
@article{huang2025distilling,
title={Distilling heterogeneous treatment effects: Stable subgroup estimation in causal inference},
author={Melody Huang and Tiffany M. Tang and Ana M. Kenney},
year={2025},
eprint={2502.07275},
archivePrefix={arXiv},
primaryClass={stat.ME},
url={https://arxiv.org/abs/2502.07275},
}These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.