
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

You can install the released version of campsismod from
CRAN with:
install.packages("campsismod")Alternatively, the package can also be installed with
devtools:
devtools::install_github("Calvagone/campsismod")Load 2-compartment PK model from built-in model library:
library(campsismod)
model <- model_suite$pk$`2cpt_fo`The model can be exported to files using write.
model %>% write(file="path_to_model_folder")In this case, the model code will be contained in the
model.campsis file. Parameters (THETA, OMEGA and SIGMA)
will be stored in their respective CSV file.
list.files("path_to_model_folder")
#> [1] "model.campsis" "omega.csv" "sigma.csv" "theta.csv"Alternatively, the model can also be exported in JSON format into a single file:
model %>% write(file="my_model.json")The model can be loaded from the previously created folder:
model <- read.campsis(file="path_to_model_folder")Or, from the previously created JSON file:
model <- read.campsis(file="my_model.json")The model can then be output in the console using
show:
show(model)
#> [MAIN]
#> TVBIO=THETA_BIO
#> TVKA=THETA_KA
#> TVVC=THETA_VC
#> TVVP=THETA_VP
#> TVQ=THETA_Q
#> TVCL=THETA_CL
#>
#> BIO=TVBIO
#> KA=TVKA * exp(ETA_KA)
#> VC=TVVC * exp(ETA_VC)
#> VP=TVVP * exp(ETA_VP)
#> Q=TVQ * exp(ETA_Q)
#> CL=TVCL * exp(ETA_CL)
#>
#> [ODE]
#> d/dt(A_ABS)=-KA*A_ABS
#> d/dt(A_CENTRAL)=KA*A_ABS + Q/VP*A_PERIPHERAL - Q/VC*A_CENTRAL - CL/VC*A_CENTRAL
#> d/dt(A_PERIPHERAL)=Q/VC*A_CENTRAL - Q/VP*A_PERIPHERAL
#>
#> [F]
#> A_ABS=BIO
#>
#> [ERROR]
#> CONC=A_CENTRAL/VC
#> if (CONC <= 0.001) CONC=0.001
#> CONC_ERR=CONC*(1 + EPS_PROP_RUV)
#>
#>
#> THETA's:
#> name index value fix label unit
#> 1 BIO 1 1 FALSE Bioavailability <NA>
#> 2 KA 2 1 FALSE Absorption rate 1/h
#> 3 VC 3 10 FALSE Volume of central compartment L
#> 4 VP 4 40 FALSE Volume of peripheral compartment L
#> 5 Q 5 20 FALSE Inter-compartment flow L/h
#> 6 CL 6 3 FALSE Clearance L/h
#> OMEGA's:
#> name index index2 value fix type
#> 1 KA 1 1 25 FALSE cv%
#> 2 VC 2 2 25 FALSE cv%
#> 3 VP 3 3 25 FALSE cv%
#> 4 Q 4 4 25 FALSE cv%
#> 5 CL 5 5 25 FALSE cv%
#> SIGMA's:
#> name index index2 value fix type
#> 1 PROP_RUV 1 1 0.1 FALSE sd
#> No variance-covariance matrix
#>
#> Compartments:
#> A_ABS (CMT=1)
#> A_CENTRAL (CMT=2)
#> A_PERIPHERAL (CMT=3)library(campsis)
dataset <- Dataset(5) %>%
add(Bolus(time=0, amount=1000, ii=12, addl=2)) %>%
add(Observations(times=0:36))
rxode <- model %>% simulate(dataset=dataset, dest="rxode2", seed=0)
mrgsolve <- model %>% simulate(dataset=dataset, dest="mrgsolve", seed=0)spaghettiPlot(rxode, "CONC")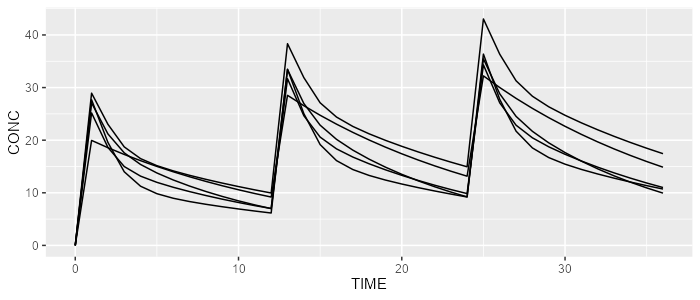
spaghettiPlot(mrgsolve, "CONC")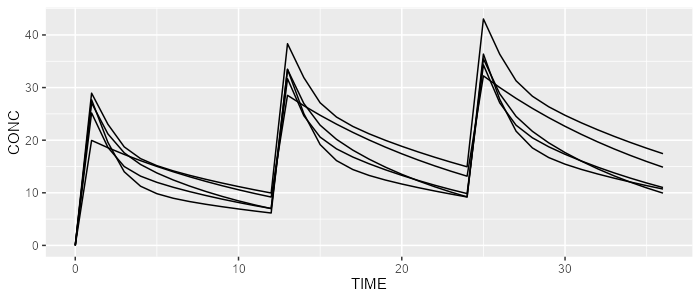
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.