
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

The goal of butterfly is to aid in the verification of continually updating timeseries data, where we expect new values over time, but want to ensure previous data remains unchanged, and timesteps remain continuous.
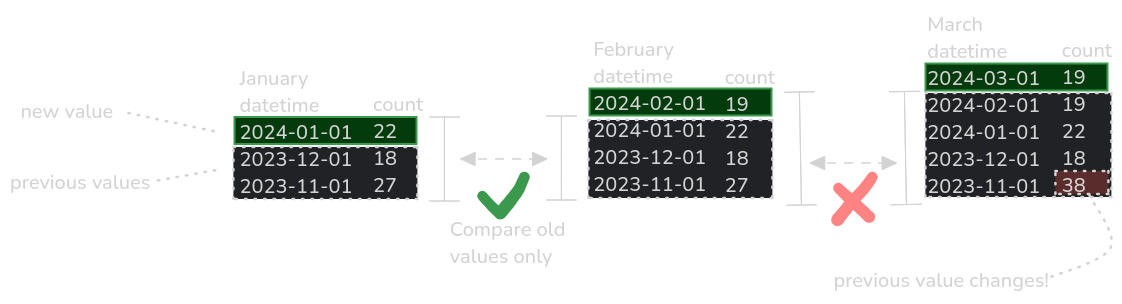
An illustration of continually updating timeseries data where a previous value unexpectedly changes.
Data previously recorded could change for a number of reasons, such as discovery of an error in model code, a change in methodology or instrument recalibration. Monitoring data sources for these changes is not always possible.
Unnoticed changes in previous data could have unintended consequences, such as invalidating a published dataset’s Digital Object Identfier (DOI), or altering future predictions if used as input in forecasting models.
Other unnoticed changes could include a jump in time or measurement frequency, due to instrument failure or software updates.
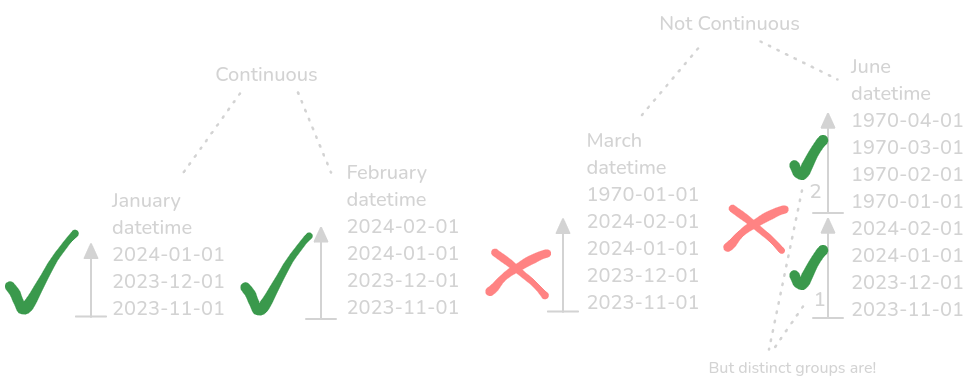
An illustration of timeseries data not being continuous in the way it is expected to be.
This package provides functionality that can be used as part of a data pipeline, to check and flag changes to previous data to prevent changes going unnoticed.
You can install butterfly with:
install.packages("butterfly", repos = "https://ropensci.r-universe.dev")The butterfly package contains the following functions:
butterfly::loupe() - examines in detail whether
previous values have changed, and returns TRUE/FALSE for no
change/change.butterfly::catch() - returns rows which contain
previously changed values in a dataframe.butterfly::release() - drops rows which contain
previously changed values, and returns a dataframe containing new and
unchanged rows.butterfly::create_object_list() - returns a list of
objects required by all of loupe(), catch()
and release(). Contains underlying functionality.butterfly::timeline() - check if a timeseries is
continuous between timesteps.butterfly::timeline_group() - group distinct, but
continuous sequences of a timeseries.There are also dummy datasets, which a fictional and purely to demonstrate butterfly functionality:
butterflycount - a list of monthly dataframes, which
contain fictional butterfly counts for a given date.forestprecipitation - a list of monthly dataframes,
which contain fictional daily precipitation measurements for a given
date.butterflymess - a messy version of
butterflycount, provided for testing purposes.This is a basic example which shows you how to use butterfly:
library(butterfly)
# Imagine a continually updated dataset that starts in January and is updated once a month
butterflycount$january
#> time count
#> 1 2024-01-01 22
#> 2 2023-12-01 55
#> 3 2023-11-01 11
# In February an additional row appears, all previous data remains the same
butterflycount$february
#> time count
#> 1 2024-02-01 17
#> 2 2024-01-01 22
#> 3 2023-12-01 55
#> 4 2023-11-01 11
# In March an additional row appears again
# ...but a previous value has unexpectedly changed
butterflycount$march
#> time count
#> 1 2024-03-01 23
#> 2 2024-02-01 17
#> 3 2024-01-01 22
#> 4 2023-12-01 55
#> 5 2023-11-01 18We can use butterfly::loupe() to examine in detail
whether previous values have changed.
butterfly::loupe(
butterflycount$february,
butterflycount$january,
datetime_variable = "time"
)
#> The following rows are new in 'df_current':
#> time count
#> 1 2024-02-01 17
#> ✔ And there are no differences with previous data.
#> [1] TRUE
butterfly::loupe(
butterflycount$march,
butterflycount$february,
datetime_variable = "time"
)
#> The following rows are new in 'df_current':
#> time count
#> 1 2024-03-01 23
#>
#> ℹ The following values have changes from the previous data.
#> old vs new
#> count
#> old[1, ] 17
#> old[2, ] 22
#> old[3, ] 55
#> - old[4, ] 18
#> + new[4, ] 11
#>
#> `old$count`: 17.0 22.0 55.0 18.0
#> `new$count`: 17.0 22.0 55.0 11.0
#> [1] FALSEbutterfly::loupe() uses dplyr::semi_join()
to match the new and old objects using a common unique identifier, which
in a timeseries will be the timestep. waldo::compare() is
then used to compare these and provide a detailed report of the
differences.
butterfly follows the waldo philosophy of
erring on the side of providing too much information, rather than too
little. It will give a detailed feedback message on the status between
two objects.
You might want to return changed rows as a dataframe, or drop them
altogether. For this butterfly::catch() and
butterfly::release() are provided.
Here, butterfly::catch() only returns rows which have
changed from the previous version. It will not return
new rows.
df_caught <- butterfly::catch(
butterflycount$march,
butterflycount$february,
datetime_variable = "time"
)
#> The following rows are new in 'df_current':
#> time count
#> 1 2024-03-01 23
#>
#> ℹ The following values have changes from the previous data.
#> old vs new
#> count
#> old[1, ] 17
#> old[2, ] 22
#> old[3, ] 55
#> - old[4, ] 18
#> + new[4, ] 11
#>
#> `old$count`: 17.0 22.0 55.0 18.0
#> `new$count`: 17.0 22.0 55.0 11.0
#>
#> ℹ Only these rows are returned.
df_caught
#> time count
#> 1 2023-11-01 18Conversely, butterfly::release() drops all rows which
had changed from the previous version. Note it retains new rows, as
these were expected.
df_released <- butterfly::release(
butterflycount$march,
butterflycount$february,
datetime_variable = "time"
)
#> The following rows are new in 'df_current':
#> time count
#> 1 2024-03-01 23
#>
#> ℹ The following values have changes from the previous data.
#> old vs new
#> count
#> old[1, ] 17
#> old[2, ] 22
#> old[3, ] 55
#> - old[4, ] 18
#> + new[4, ] 11
#>
#> `old$count`: 17.0 22.0 55.0 18.0
#> `new$count`: 17.0 22.0 55.0 11.0
#>
#> ℹ These will be dropped, but new rows are included.
df_released
#> time count
#> 1 2024-03-01 23
#> 2 2024-02-01 17
#> 3 2024-01-01 22
#> 4 2023-12-01 55timeline()To check if a timeseries is continuous, timeline() and
timeline_group() are provided.
# A rain gauge which measures precipitation every day
butterfly::forestprecipitation$january
#> time rainfall_mm
#> 1 2024-01-01 0.0
#> 2 2024-01-02 2.6
#> 3 2024-01-03 0.0
#> 4 2024-01-04 0.0
#> 5 2024-01-05 3.7
#> 6 2024-01-06 0.8
# In February there is a power failure in the instrument
butterfly::forestprecipitation$february
#> time rainfall_mm
#> 1 2024-02-01 1.1
#> 2 2024-02-02 0.0
#> 3 2024-02-03 1.4
#> 4 2024-02-04 2.2
#> 5 1970-01-01 3.4
#> 6 1970-01-02 0.6To check if a timeseries is continuous:
butterfly::timeline(
forestprecipitation$january,
datetime_variable = "time",
expected_lag = 1
)
#> ✔ There are no time lags which are greater than the expected lag: 1 days. By this measure, the timeseries is continuous.
#> [1] TRUEIn February our imaginary rain gauge’s onboard computer had a failure.
The timestamp was reset to 1970-01-01:
forestprecipitation$february
#> time rainfall_mm
#> 1 2024-02-01 1.1
#> 2 2024-02-02 0.0
#> 3 2024-02-03 1.4
#> 4 2024-02-04 2.2
#> 5 1970-01-01 3.4
#> 6 1970-01-02 0.6
butterfly::timeline(
forestprecipitation$february,
datetime_variable = "time",
expected_lag = 1
)
#> ℹ There are time lags which are greater than the expected lag: 1 days. This indicates the timeseries is not continuous. There are 2 distinct continuous sequences. Use `timeline_group()` to extract.
#> [1] FALSEIf we wanted to group chunks of our timeseries that are distinct, or
broken up in some way, but still continuous, we can use
timeline_group():
butterfly::timeline_group(
forestprecipitation$february,
datetime_variable = "time",
expected_lag = 1
)
#> time rainfall_mm timelag timeline_group
#> 1 2024-02-01 1.1 NA days 1
#> 2 2024-02-02 0.0 1.00 days 1
#> 3 2024-02-03 1.4 1.00 days 1
#> 4 2024-02-04 2.2 1.00 days 1
#> 5 1970-01-01 3.4 -19757.04 days 2
#> 6 1970-01-02 0.6 1.00 days 2The butterfly package was created for a specific use case of handling continuously updating/overwritten timeseries data, where previous values may change without notice.
There are other R packages and functions which handle object
comparison, which may suit your specific needs better. Below we describe
their overlap and differences to butterfly:
butterfly uses waldo::compare() in every
function to provide a report on difference. There is therefore
significant overlap, however butterfly builds on
waldo by providing the functionality of comparing objects
where we expect some changes, with previous versions but not others.
butterfly also provides extra user feedback to provide
clarity on what it is and isn’t comparing, due to the nature of
comparing only “matched” rows.waldo, but specifically for data frames,
diffdf provides the ability to compare data frames
directly. We could have used diffdf::diffdf() in our case,
but we prefer waldo’s more explicit and clear user
feedback. That said, there is significant overlap in functionality:
butterfly::loupe() and
diffdf::diffdf_has_issues() both provide a TRUE/FALSE
difference check, while diffdf::diffdf_issue_rows() and
butterfly::catch() both return the rows where changes have
occurred. However, it lacks the flexibility of butterfly to
compare object where we expect some changes, but not others.assertr provides assertion functionality that can be used
as part of a pipeline, and test assertions on a particular dataset, but
it does not offer tools for comparison. We do highly recommend using
assertr for checks, prior to using butterfly,
as any data quality issues will be caught first.daquiri provides tools to check data quality and visually
inspect timeseries data. It is also quality assurance package for
timeseries, but has a very different purpose to
butterfly.Other functions include all.equal() (base R) or dplyr’s
setdiff().
butterfly in
productionRead more about how butterfly is used
in an operational data pipeline to verify a continually updated
and published dataset.
For full guidance on contributions, please refer to
.github/CONTRIBUTING.md.
Corrections, suggestions and general improvements are welcome as issues.
You can also suggest changes by forking this repository, and opening a pull request. Please target your pull requests to the main branch.
You can push directly to main for small fixes. Please use PRs to main for discussing larger updates.
Please note that this package is released with a Contributor Code of Conduct.
By contributing to this project, you agree to abide by its terms.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.