The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
anndata
is a commonly used Python package for keeping track of data and learned
annotations, and can be used to read from and write to the h5ad file
format. It is also the main data format used in the scanpy python
package (Wolf, Angerer, and Theis 2018).
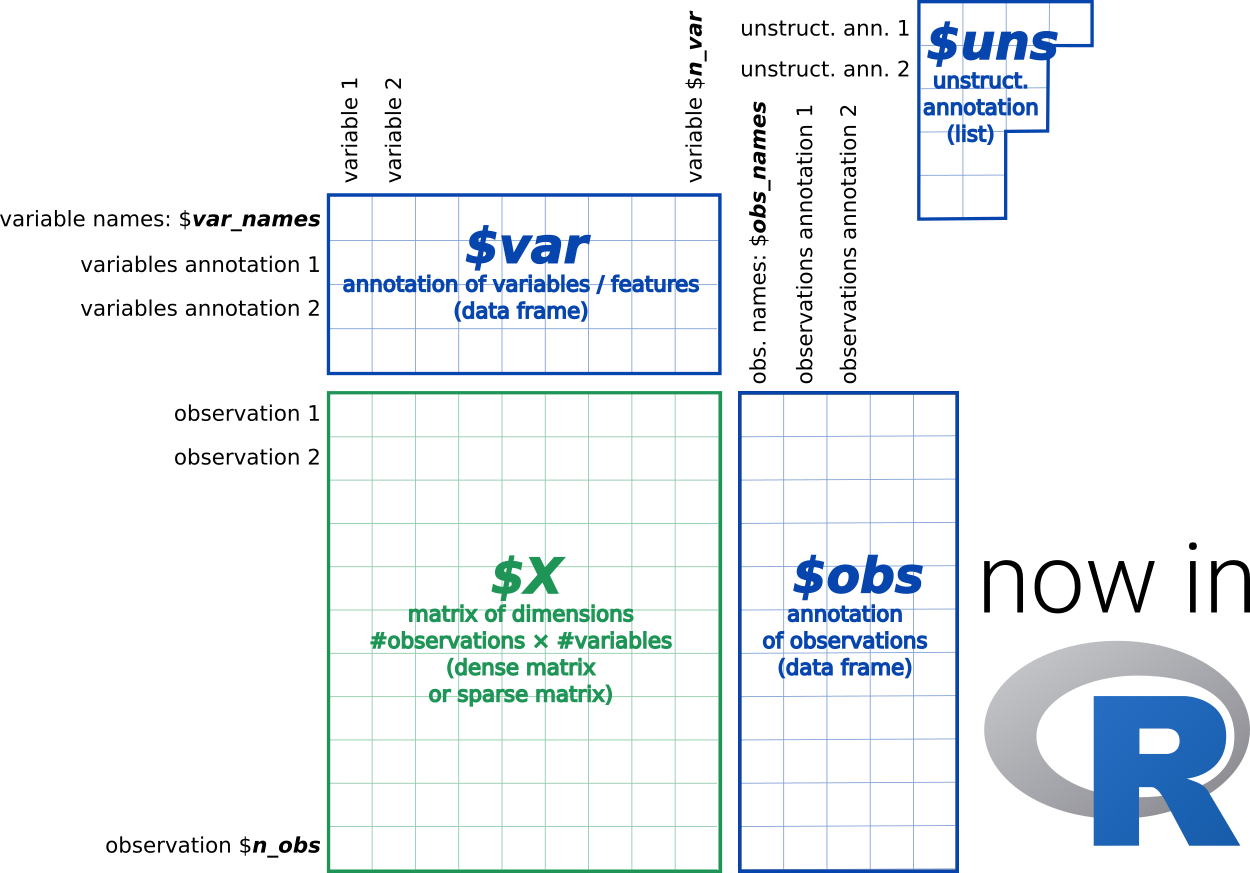
However, using scanpy/anndata in R can be a major hassle. When trying to read an h5ad file, R users could approach this problem in one of two ways. A) You could read in the file manually (since it’s an H5 file), but this involves a lot of manual work and a lot of understanding on how the h5ad and H5 file formats work (also, expect major headaches from cryptic hdf5r bugs). Or B) interact with scanpy and anndata through reticulate, but run into issues converting some of the python objects into R.
We recently published anndata
on CRAN, which is a reticulate wrapper for the Python package – with
some syntax sprinkled on top to make R users feel more at home.
anndata for R is still under active development at github.com/dynverse/anndata. If you encounter any issues, feel free to post an issue on GitHub!
You can install anndata for R from CRAN as follows:
install.packages("anndata")Normally, reticulate should take care of installing Miniconda and the Python anndata.
If not, try running:
reticulate::install_miniconda()
anndata::install_anndata()The API of anndata for R is very similar to its Python counterpart. Here is an example:
library(anndata)##
## Attaching package: 'anndata'
## The following object is masked from 'package:readr':
##
## read_csvad <- read_h5ad("example_formats/pbmc_1k_protein_v3_processed.h5ad")
ad## AnnData object with n_obs × n_vars = 713 × 33538
## var: 'gene_ids', 'feature_types', 'genome', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
## uns: 'hvgParameters', 'normalizationParameters', 'pca', 'pcaParameters'
## obsm: 'X_pca'
## varm: 'PCs'Matrix::rowMeans(ad$X[1:10,])## AAACCCAAGTGGTCAG-1 AAAGGTATCAACTACG-1 AAAGTCCAGCGTGTCC-1 AACACACTCAAGAGTA-1
## 0.06499579 0.06385104 0.06102355 0.06739055
## AACACACTCGACGAGA-1 AACAGGGCAGGAGGTT-1 AACAGGGCAGTGTATC-1 AACAGGGTCAGAATAG-1
## 0.08891241 0.08648681 0.09318970 0.09140243
## AACCTGAAGATGGTCG-1 AACGGGATCGTTATCT-1
## 0.06664118 0.07866523See ?anndata for a full list of the functions provided
by this package. Check out any of the other vignettes by clicking any of
the links below:
In some cases, this package may still act more like a Python package
rather than an R package. Some more helper functions and helper classes
need to be defined in order to fully encapsulate AnnData()
objects. Examples are ad$chunked_X(...), backed file modes,
read_zarr() and ad$write_zarr().
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.