The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This package is an implementation of the additive profile clustering (ADPROCLUS) method in R. It can be used to obtain overlapping clustering models for object-by-variable data matrices. It also contains the low dimensional ADPROCLUS method, which achieves a simultaneous dimension reduction when searching for overlapping clusters. This can be used when the object-by-variable data contains a very large number of variables.
You can install the latest version from CRAN:
install.packages("adproclus")Or install the development version of ADPROCLUS from GitHub with:
# install.packages("devtools")
devtools::install_github("henry-heppe/adproclus")This is a basic example which shows you how to use the regular ADPROCLUS and the low dimensional ADPROCLUS:
library("adproclus")
# import data
our_data <- adproclus::CGdata
# perform ADPROCLUS to get an overlapping clustering model
model_full <- adproclus(data = our_data, nclusters = 2)
# perform low dimensional ADPROCLUS to get an overlapping clustering model in terms of a smaller number of variables
model_lowdim <- adproclus_low_dim(data = our_data, nclusters = 3, ncomponents = 2)To select the number of clusters (and the number of components in the low dimensional case) the package provides two model selection functions.
library("adproclus")
# estimate multiple ADPROCLUS models
models <- mselect_adproclus(data = CGdata, min_nclusters = 2, max_nclusters = 4)
# estimate multiple low dimensional ADPROCLUS models
models_lowdim <- mselect_adproclus_low_dim(data = CGdata, min_nclusters = 2, max_nclusters = 4, min_ncomponents = 1, max_ncomponents = 3)
# visualize models as a scree plot
plot_scree_adpc(models)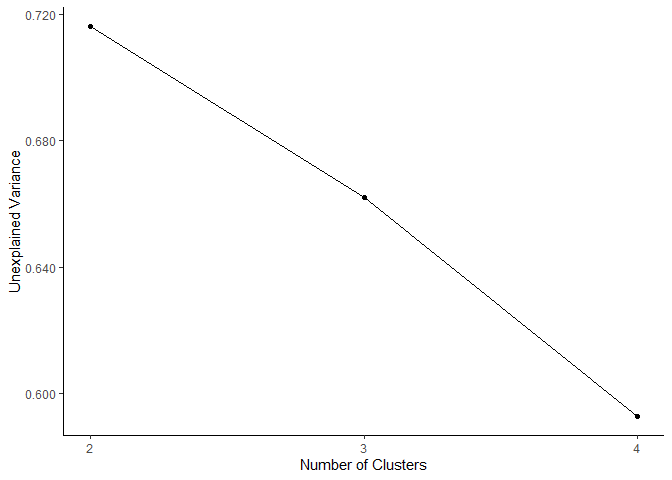
# visualize the low dimensional models as a scree plot
plot_scree_adpc(models_lowdim)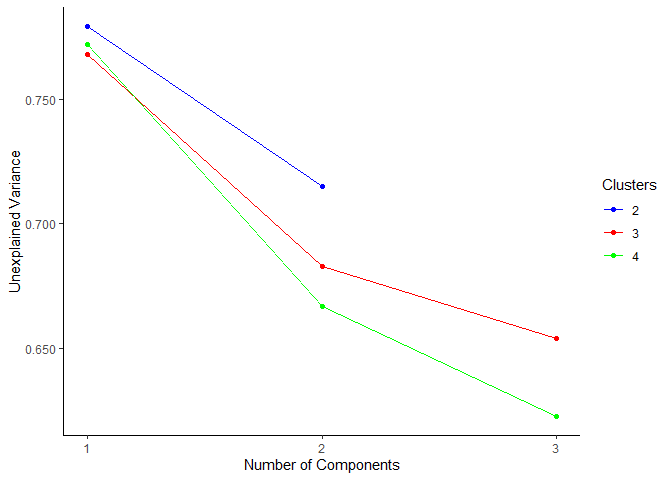
# select the best full dimensional model
best_model <- select_by_CHull(models)
# select a the conditionally optimal low dimensional model for each number of clusters
best_models_lowdim <- select_by_CHull(models_lowdim)
# visualize the preselected set of low dimensional models
plot_scree_adpc_preselected(best_models_lowdim)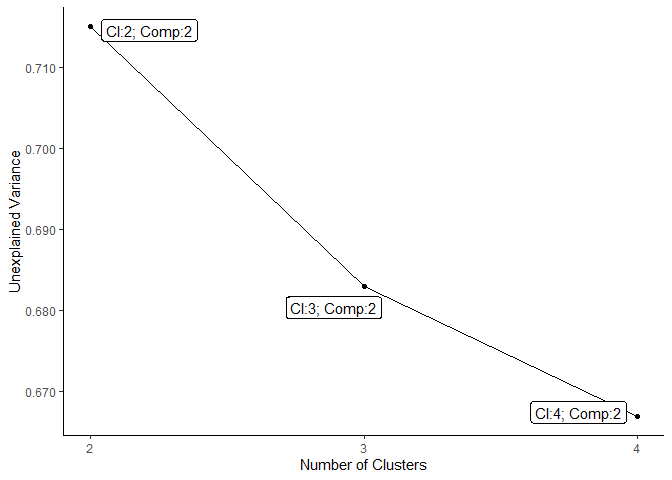
The package also provides functionality to obtain membership matrices, which the algorithm can start the alternating least squares procedure on. There are three different possibilities to obtain such matrices: random, semi-random and rational (see respective function documentation for details).
library("adproclus")
# import data
our_data <- adproclus::CGdata
# Obtaining a membership matrix were the entries are randomly assigned values of 0 or 1
start_allocation1 <- get_random(our_data, 3)
# Obtaining a membership matrix based on a profile matrix consisting of randomly selected rows of the data
start_allocation2 <- get_semirandom(our_data, 3)
# Obtaining a user-defined rational start profile matrix (here the first 3 rows of the data)
start_allocation3 <- get_rational(our_data, our_data[1:3, ])$AThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.