
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

Chest pain? Calculate your cardiovascular risk score.
The goal of RiskScorescvd R package is to calculate the most commonly used cardiovascular risk scores. Original research publication can be found here: https://openheart.bmj.com/content/11/2/e002755
We have developed nine of the most commonly used risk scores with a dependency (ASCVD [PooledCohort]) making the following available:
NOTE: Troponin I values should be used. Additional functions for Troponin T are under development
You can install from CRAN with:
# Install from CRAN
install.packages("RiskScorescvd")You can install the development version of RiskScorescvd from GitHub with:
# install.packages("devtools")
devtools::install_github("dvicencio/RiskScorescvd")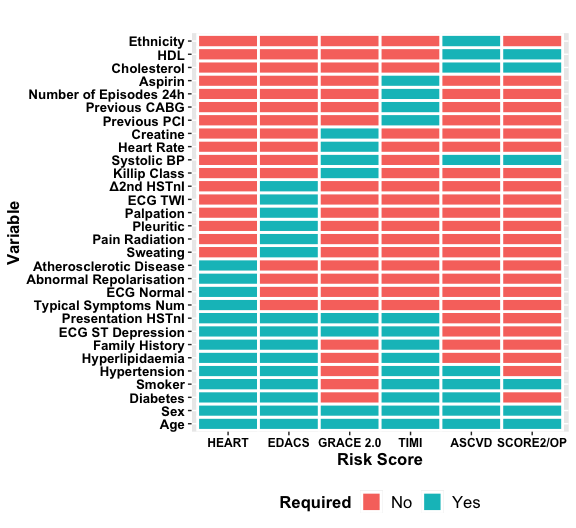
This is a basic example of how the data set should look to calculate all risk scores available in the package:
library(RiskScorescvd)
#> Loading required package: PooledCohort
# Create a data frame or list with the necessary variables
# Set the number of rows
num_rows <- 100
# Create a large dataset with 100 rows
cohort_xx <- data.frame(
typical_symptoms.num = as.numeric(sample(0:6, num_rows, replace = TRUE)),
ecg.normal = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
abn.repolarisation = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
ecg.st.depression = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
Age = as.numeric(sample(30:80, num_rows, replace = TRUE)),
diabetes = sample(c(1, 0), num_rows, replace = TRUE),
smoker = sample(c(1, 0), num_rows, replace = TRUE),
hypertension = sample(c(1, 0), num_rows, replace = TRUE),
hyperlipidaemia = sample(c(1, 0), num_rows, replace = TRUE),
family.history = sample(c(1, 0), num_rows, replace = TRUE),
atherosclerotic.disease = sample(c(1, 0), num_rows, replace = TRUE),
presentation_hstni = as.numeric(sample(10:100, num_rows, replace = TRUE)),
Gender = sample(c("male", "female"), num_rows, replace = TRUE),
sweating = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
pain.radiation = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
pleuritic = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
palpation = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
ecg.twi = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
second_hstni = as.numeric(sample(1:200, num_rows, replace = TRUE)),
killip.class = as.numeric(sample(1:4, num_rows, replace = TRUE)),
heart.rate = as.numeric(sample(0:300, num_rows, replace = TRUE)),
systolic.bp = as.numeric(sample(40:300, num_rows, replace = TRUE)),
aspirin = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
creat = as.numeric(sample(0:4, num_rows, replace = TRUE)),
number.of.episodes.24h = as.numeric(sample(0:20, num_rows, replace = TRUE)),
previous.pci = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
cardiac.arrest = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
previous.cabg = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
total.chol = as.numeric(sample(5:100, num_rows, replace = TRUE)),
total.hdl = as.numeric(sample(2:5, num_rows, replace = TRUE)),
Ethnicity = sample(c("white", "black", "asian", "other"), num_rows, replace = TRUE),
eGFR = as.numeric(sample(15:120, num_rows, replace = TRUE)),
ACR = as.numeric(sample(5:1500, num_rows, replace = TRUE)),
trace = sample(c("trace", "1+", "2+", "3+", "4+"), num_rows, replace = TRUE)
)
str(cohort_xx)
#> 'data.frame': 100 obs. of 34 variables:
#> $ typical_symptoms.num : num 1 1 0 2 4 6 6 2 4 4 ...
#> $ ecg.normal : num 0 1 1 0 0 0 0 1 1 0 ...
#> $ abn.repolarisation : num 1 0 1 1 0 0 0 1 0 1 ...
#> $ ecg.st.depression : num 0 1 0 1 1 0 1 0 1 0 ...
#> $ Age : num 61 75 60 44 47 46 71 39 56 50 ...
#> $ diabetes : num 1 1 1 1 0 1 0 0 1 1 ...
#> $ smoker : num 0 1 0 1 0 0 1 1 1 1 ...
#> $ hypertension : num 1 1 1 1 1 0 1 0 0 0 ...
#> $ hyperlipidaemia : num 1 0 1 0 1 0 0 1 1 0 ...
#> $ family.history : num 1 0 0 0 1 1 0 1 0 1 ...
#> $ atherosclerotic.disease: num 1 1 1 0 1 0 0 1 1 0 ...
#> $ presentation_hstni : num 19 75 96 76 82 35 36 20 54 27 ...
#> $ Gender : chr "male" "female" "male" "female" ...
#> $ sweating : num 1 0 0 1 1 0 1 0 1 0 ...
#> $ pain.radiation : num 1 1 1 1 1 1 1 1 0 1 ...
#> $ pleuritic : num 1 1 0 0 0 1 1 1 0 0 ...
#> $ palpation : num 1 0 1 0 0 0 0 0 0 1 ...
#> $ ecg.twi : num 0 1 1 0 0 1 1 0 0 1 ...
#> $ second_hstni : num 157 131 58 45 174 23 178 189 20 78 ...
#> $ killip.class : num 3 4 4 3 2 3 2 2 1 4 ...
#> $ heart.rate : num 227 273 122 169 147 291 267 266 198 148 ...
#> $ systolic.bp : num 41 218 153 118 282 92 191 293 101 267 ...
#> $ aspirin : num 1 0 0 1 0 0 1 0 1 1 ...
#> $ creat : num 3 3 2 3 3 0 1 2 4 3 ...
#> $ number.of.episodes.24h : num 11 12 19 18 12 17 10 16 4 8 ...
#> $ previous.pci : num 0 0 0 1 0 1 0 1 1 0 ...
#> $ cardiac.arrest : num 0 1 0 0 1 1 0 0 1 1 ...
#> $ previous.cabg : num 0 1 1 0 0 0 0 1 0 0 ...
#> $ total.chol : num 97 89 52 93 29 56 58 99 91 40 ...
#> $ total.hdl : num 3 3 4 3 4 5 5 4 5 2 ...
#> $ Ethnicity : chr "other" "white" "white" "white" ...
#> $ eGFR : num 77 69 110 81 22 76 68 58 43 65 ...
#> $ ACR : num 782 1161 1005 595 840 ...
#> $ trace : chr "trace" "4+" "4+" "trace" ...This is a basic example of how to calculate all risk scores available in the package and create a new data set with 12 new variables of the calculated and classified risk scores:
# Call the function with the cohort_xx to calculate all risk scores available in the package
new_data_frame <- calc_scores(data = cohort_xx)
# Select columns created after calculation
All_scores <- new_data_frame %>% select(HEART_score, HEART_strat, EDACS_score, EDACS_strat, GRACE_score, GRACE_strat, TIMI_score, TIMI_strat, SCORE2_score, SCORE2_strat, ASCVD_score, ASCVD_strat)
# Observe the results
head(All_scores)
#> # A tibble: 6 × 12
#> # Rowwise:
#> HEART_score HEART_strat EDACS_score EDACS_strat GRACE_score GRACE_strat
#> <dbl> <ord> <dbl> <ord> <dbl> <ord>
#> 1 4 Moderate risk 14 Not low risk 150 High risk
#> 2 8 High risk 15 Not low risk 181 High risk
#> 3 5 Moderate risk 13 Not low risk 132 High risk
#> 4 7 High risk 14 Not low risk 131 High risk
#> 5 8 High risk 16 Not low risk 88 Low risk
#> 6 6 Moderate risk 5 Not low risk 132 High risk
#> # ℹ 6 more variables: TIMI_score <dbl>, TIMI_strat <ord>, SCORE2_score <dbl>,
#> # SCORE2_strat <ord>, ASCVD_score <dbl>, ASCVD_strat <ord>
# Create a summary of them to obtain an initial idea of distribution
summary(All_scores)
#> HEART_score HEART_strat EDACS_score EDACS_strat
#> Min. : 2.00 Low risk : 6 Min. :-2.00 Low risk : 1
#> 1st Qu.: 5.00 Moderate risk:54 1st Qu.: 5.00 Not low risk:99
#> Median : 6.00 High risk :40 Median :10.00
#> Mean : 6.06 Mean :10.78
#> 3rd Qu.: 7.00 3rd Qu.:16.00
#> Max. :10.00 Max. :30.00
#> GRACE_score GRACE_strat TIMI_score TIMI_strat
#> Min. : 29.00 Low risk :30 Min. :2.00 Very low risk: 0
#> 1st Qu.: 85.75 Moderate risk:34 1st Qu.:4.00 Low risk : 6
#> Median :107.00 High risk :36 Median :4.00 Moderate risk:49
#> Mean :108.53 Mean :4.44 High risk :45
#> 3rd Qu.:132.00 3rd Qu.:5.00
#> Max. :205.00 Max. :7.00
#> SCORE2_score SCORE2_strat ASCVD_score ASCVD_strat
#> Min. : 0.00 Very low risk: 0 Min. :0.0000 Very low risk: 7
#> 1st Qu.: 55.00 Low risk : 6 1st Qu.:0.1900 Low risk : 2
#> Median :100.00 Moderate risk: 6 Median :0.4650 Moderate risk:17
#> Mean : 75.87 High risk :88 Mean :0.5027 High risk :74
#> 3rd Qu.:100.00 3rd Qu.:0.8300
#> Max. :100.00 Max. :1.0000These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.