
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
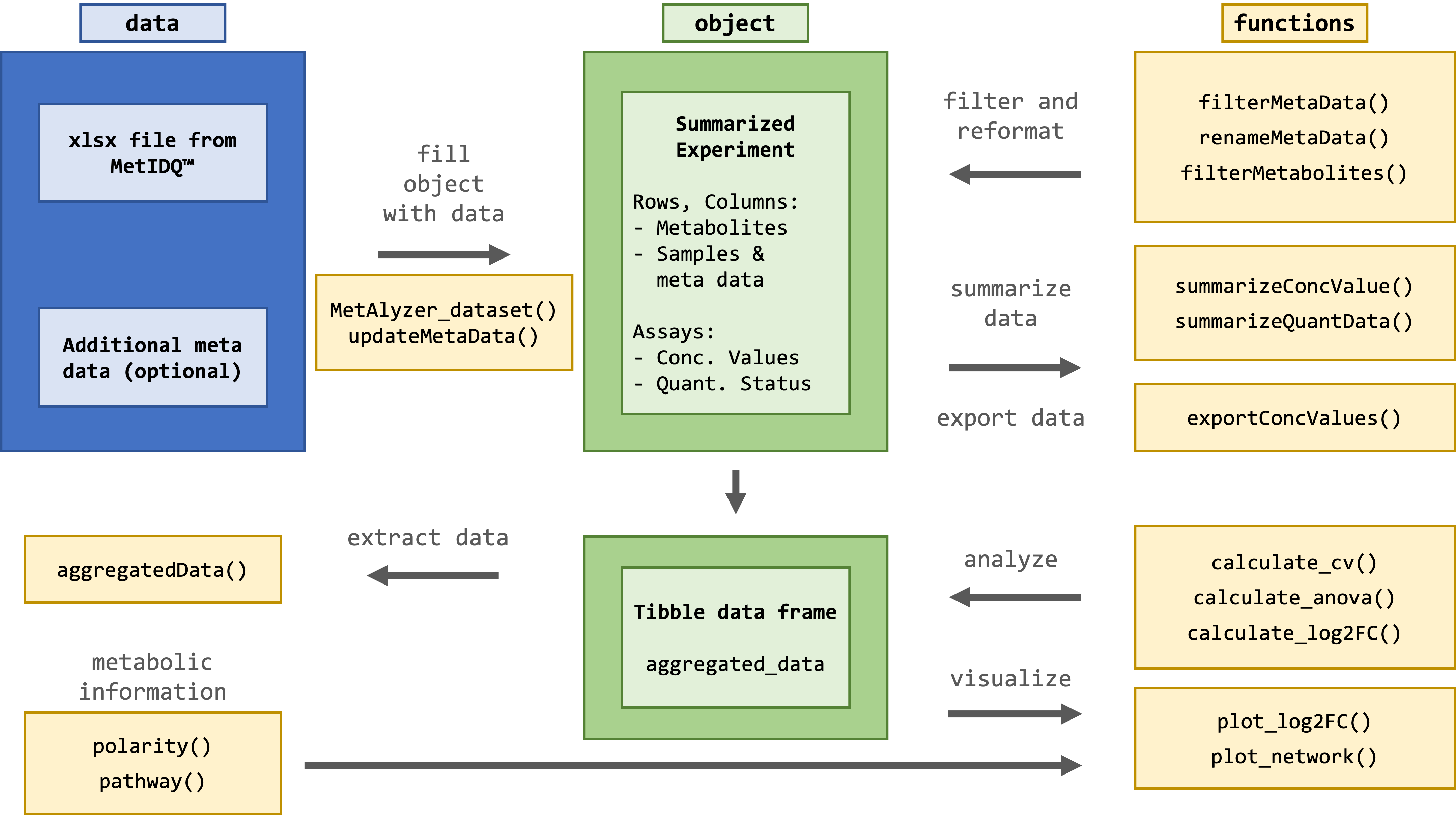
An R Package to read and analyze MetIDQ™ output
The package provides methods to read output files from the MetIDQ™ software into R. Metabolomics data is read and reformatted into an S4 object for convenient data handling, statistics and downstream analysis.
There is a version available on CRAN.
install.packages("MetAlyzer")For the latest version install from GitHub
library(devtools)
install_github("nilsmechtel/MetAlyzer")The package takes metabolomic measurements and the quantification status (e.g. “Valid”, “LOQ”, “LOD”) as “.xlsx” files generated from the MetIDQ™ software. Additionally, meta data for each sample can be provided for further analysis.
This is an extract from one of the provided example data sets. 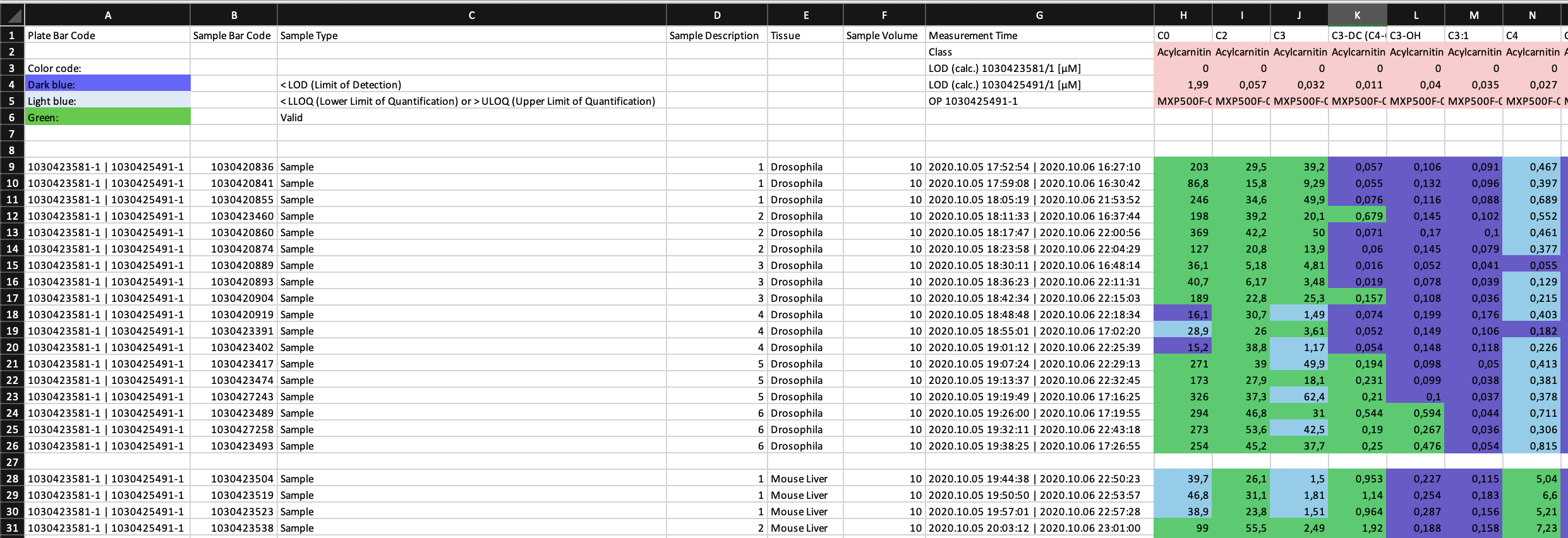
> metalyzer_se <- MetAlyzer_dataset(file_path = extraction_data())
_____ ______ _______ _________ ________ ___ ___ ___ ________ _______ ________
|\ _ \ _ \|\ ___ \|\___ ___\\ __ \|\ \ |\ \ / /|\_____ \|\ ___ \ |\ __ \
\ \ \\\__\ \ \ \ __/\|___ \ \_\ \ \|\ \ \ \ \ \ \/ / /\|___/ /\ \ __/|\ \ \|\ \
\ \ \\|__| \ \ \ \_|/__ \ \ \ \ \ __ \ \ \ \ \ / / / / /\ \ \_|/_\ \ _ _\
\ \ \ \ \ \ \ \_|\ \ \ \ \ \ \ \ \ \ \ \____ \/ / / / /_/__\ \ \_|\ \ \ \\ \|
\ \__\ \ \__\ \_______\ \ \__\ \ \__\ \__\ \_______\__/ / / |\________\ \_______\ \__\\ _\
\|__| \|__|\|_______| \|__| \|__|\|__|\|_______|\____/ / \|_______|\|_______|\|__|\|__|
\|____|/
Info: Reading color code "FFFFCCCC" as "#FFCCCC"
Info: Reading color code "FF00CD66" as "#00CD66"
Info: Reading color code "FF6A5ACD" as "#6A5ACD"
Info: Reading color code "FF87CEEB" as "#87CEEB"
Info: Reading color code "FFFFFFCC" as "#FFFFCC"
Measured concentration values:
------------------------------
0% 25% 50% 75% 100%
0.000 0.017 1.760 21.200 288149.000
NAs: 5348 (8.38%)
Note: 'Metabolism Indicators' are frequently NA!
Measured quantification status:
-------------------------------
Valid: 24095 (37.77%)
LOQ: 5799 (9.09%)
LOD: 21789 (34.16%)
Invalid: 12105 (18.98%)
NAs: 0 (0%)For further filtering, statistical analysis and plotting, the data is reformatted and aggregated into a tibble data frame.
> aggregatedData(metalyzer_se)
# A tibble: 63,788 × 5
# Groups: Metabolite [862]
ID Metabolite Class Concentration Status
<fct> <fct> <fct> <dbl> <fct>
1 9 C0 Acylcarnitines 203 Valid
2 10 C0 Acylcarnitines 86.8 Valid
3 11 C0 Acylcarnitines 246 Valid
4 12 C0 Acylcarnitines 198 Valid
5 13 C0 Acylcarnitines 369 Valid
6 14 C0 Acylcarnitines 127 Valid
7 15 C0 Acylcarnitines 36.1 Valid
8 16 C0 Acylcarnitines 40.7 Valid
9 17 C0 Acylcarnitines 189 Valid
10 18 C0 Acylcarnitines 16.1 LOD
# ℹ 63,778 more rows
# ℹ Use `print(n = ...)` to see more rowsFor a comprehensive tutorial, please check out the MetAlyzer Vignette.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.