
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

The MedLEA package provides morphological and structural features of 471 medicinal plant leaves and 1099 leaf images of 31 species and 29-45 images per species.
You could install the stable version on CRAN:
install.packages("MedLEA")You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("SMART-Research/MedLEA")



library(MedLEA)
data("medlea")
head(medlea)
#> ID Sinhala_Name Family_Name
#> 1 1 Tel kaduru (???? ?????) EUPHORBIACEAE
#> 2 2 Telhiriya (?????????) / Mayura manikkam (???? ?????????) RHAMNACEAE
#> 3 3 Thakkali SOLANACEAE
#> 4 4 Thala PEDALIACEAE
#> 5 5 Thana hal POACEAE
#> 6 6 Thebu (????) / Koltan (???????) ZINGIBERACEAE
#> Scientific_Name Shape Arrangements Bipinnately_compound
#> 1 Sapium insigne Round Simple False
#> 2 Colubrina asiatica var. asiatica Round Simple False
#> 3 Lycopersicon esculentum Diamond Compound False
#> 4 Sesamum indicum Diamond Simple False
#> 5 Setaria italica Diamond Simple False
#> 6 Costus speciosus Round Simple False
#> Pinnately_compound Palmately_compound Edges Uniform_background Red_Margin
#> 1 False False Smooth True False
#> 2 False False Toothed True False
#> 3 True False Lobed True False
#> 4 False False Smooth True False
#> 5 False False Smooth True False
#> 6 False False Smooth True False
#> Shaded_margin White_Shading Red_Shading White_line Green_leaf Red_leaf
#> 1 False False False False True False
#> 2 False False False False True False
#> 3 False False False False True False
#> 4 False True False False True False
#> 5 False False False False True False
#> 6 False False False False True False
#> Veins Arrangement_on_the_stem Leaf_Apices Leaf_Base
#> 1 Pinnate Whorled Acute Obtuse
#> 2 Pinnate Alternate Acute Acuate
#> 3 Pinnate Opposite Obtuse Cordate
#> 4 Pinnate Whorled Acute Cuneate
#> 5 Parallel Opposite Acute Gradually tapering
#> 6 Parallel Alternate Acute Obtuselibrary(ggplot2)
library(wordcloud2)
library(magrittr)
library(patchwork)
library(dplyr)
library(tm)
#unique(medlea$Family_Name)
text1 <- medlea$Family_Name
docs <- Corpus(VectorSource(text1))
docs <- docs%>% tm_map(stripWhitespace)
dtm <- TermDocumentMatrix(docs)
matrix <- as.matrix(dtm)
words <- sort(rowSums(matrix), decreasing = TRUE)
df <- data.frame(word = names(words), freq = words)
p1 <- wordcloud2(data = df, size = 0.9,color = 'random-dark', shape = 'pentagon')
p1
medlea <- filter(medlea, Arrangements == "Simple")
d11 <- as.data.frame(table(medlea$Shape))
names(d11) <- c('Shape_of_the_leaf', 'No_of_leaves')
p2 <- ggplot(d11, aes(x= reorder(Shape_of_the_leaf, No_of_leaves), y=No_of_leaves)) + labs(y="Number of leaves", x="Shape of the leaf") + geom_bar(stat = "identity", width = 0.6) + ggtitle("Composition of the Sample by the Shape Label") + coord_flip()d11 <- as.data.frame(table(medlea$Edges))
names(d11) <- c('Edges', 'No_of_leaves')
#d11 <- d11 %>% mutate(Percentage = round(No_of_leaves*100/sum(No_of_leaves),0))
#ggplot(d11, aes(x= reorder(Shape_of_the_leaf, Percentage), y=Percentage)) + labs(y="Percentage", x="Shape of the leaf") + geom_bar(stat = "identity", width = 0.5) + geom_label(aes(label = paste0(Percentage, "%")), nudge_y = -3, size = 3.25, label.padding = unit(0.175,"lines")) + ggtitle("Composition of the Sample by the Shape Label") + coord_flip()
p3 <- ggplot(d11, aes(x= reorder(Edges, No_of_leaves), y=No_of_leaves)) + labs(y="Number of leaves", x="Edge type of the leaf") + geom_bar(stat = "identity", width = 0.6) + ggtitle("Composition of the Sample by the Edge Type") + coord_flip()
p2 + p3 + plot_layout(ncol = 1)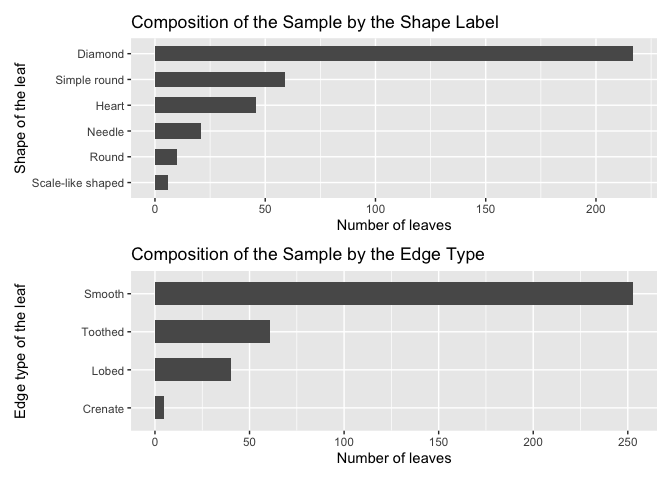
medlea <- filter(medlea, Shape != "Scale-like shaped")
d29 <- as.data.frame(table(medlea$Shape,medlea$Edges))
names(d29) <- c('Shape','Edges','No_of_leaves')
d29$Edges <- factor(d29$Edges, levels = c("Smooth", "Toothed","Lobed","Crenate"))
ggplot(d29, aes(fill = Edges, x=Shape , y=No_of_leaves)) + labs(y="Number of leaves", x="Shape of the leaf") + geom_bar(stat = "identity", width = 0.5, position = position_dodge()) + coord_flip() + ggtitle("Composition of the sample by Shape Label and Edge type") + scale_fill_brewer(palette = "Set1") 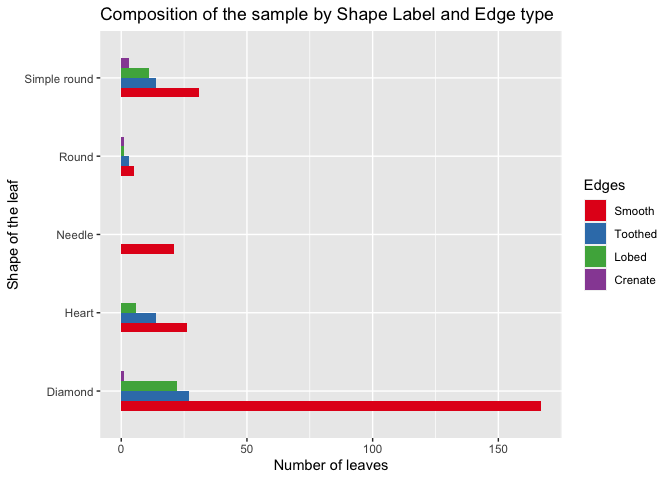
load_images()
[1] "The repository of leaf images of medicinal plants in Sri Lanka is collected by following the image acquisition steps that we identified."
[1] "The repository contains 1099 leaf images of 31 species and 29-45 images per species.These have simple arrangement. The photographs were taken from the device, Huawei nova 3i. The closest photographs were captured on a white background."
[1] "All the leaf images are in a google drive folder that anyone can access. You can download the images directly from the drive."
[1] "The shareable link: https://drive.google.com/drive/folders/1W3ap8UhBCIVN5U_UUVSZeTh7VG4Jqbev?usp=sharing"These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.