The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The goal of MBNMAdose is to provide a collection of
useful commands that allow users to run dose-response Model-Based
Network Meta-Analyses (MBNMA). This allows evidence synthesis of studies
that compare multiple doses of different agents in a way that can
account for the dose-response relationship.
Whilst making use of all the available evidence in a statistically robust and biologically plausible framework, this also can help connect networks at the agent level that may otherwise be disconnected at the dose/treatment level, and help improve precision of estimates(Pedder et al. 2021). It avoids “lumping” of doses that is often done in standard Network Meta-Analysis (NMA). All models and analyses are implemented in a Bayesian framework, following an extension of the standard NMA methodology presented by (Lu and Ades 2004) and are run in JAGS (Just Another Gibbs Sampler). For full details of dose-response MBNMA methodology see Mawdsley et al. (2016). Throughout this package we refer to a treatment as a specific dose or a specific agent.
A short introductory YouTube video from the ESMAR Conference 2021 can be found here
On CRAN you can easily install the current release version of
MBNMAdose from CRAN with:
install.packages("MBNMAdose")For the development version the package can be installed directly
from GitHub using the devtools R package:
# First install devtools
install.packages("devtools")
# Then install MBNMAdose directly from GitHub
devtools::install_github("hugaped/MBNMAdose")Functions within MBNMAdose follow a clear pattern of
use:
mbnma.network() and explore potential relationshipsmbnma.run().
Modelling of effect modifying covariates is also possibly using Network
Meta-Regression.nma.nodesplit() and nma.run()predict()At each of these stages there are a number of informative plots that can be generated to help understand the data and to make decisions regarding model fitting. Exported functions in the package are connected like so:
MBNMAdose package structure: Light green nodes represent classes
and the generic functions that can be applied to them. Dashed boxes
indicate functions that can be applied to objects of specific
classes 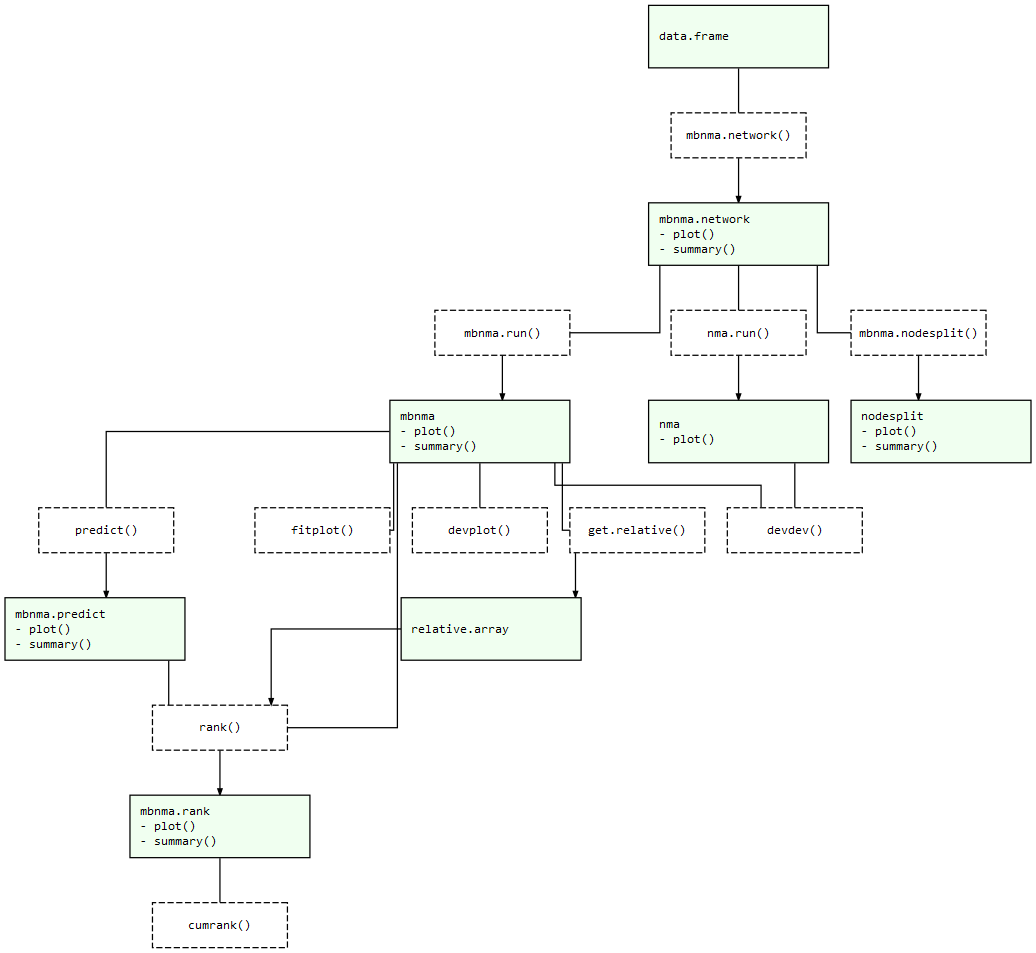
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.