The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This package provides functions to replicate the CDC’s LTAS program within R. Currently, it will only stratify a cohort for fixed strata. It does not consider a history file for time-dependent exposures/variables.
It comes pre-installed with referent underlying cause of death (UCOD) rates for the 119 minors defined in Minors119 vignette for the US population.
You can install LTASR like so:
install.packages('LTASR')It is highly recommended to use R as well as RStudio. RStudio is a separate piece of software used to write and run R code.
install.packages('tidyverse')This will only need to be run once, ever. It will install the necessary packages and functions to your computer.
| Variable | Description | Format |
|---|---|---|
| id | Unique identifier for each person | |
| gender | Sex of person (“M” = male / “F” = female) | numeric |
| race | Race of person (“W” = white / “N” = nonwhite) | numeric |
| dob | date of birth | character |
| pybegin | date to begin follow-up | character |
| dlo | date last observed. Minimum of end of study, date of death, date lost to follow-up | character |
| vs | an indicator of ‘D’ for those who are deceased. All other values will be treated as alive or censored as of dlo. | character |
| code | ICD code of death; missing if person is censored or alive at study end date | character |
| rev | revision of ICD code (7-10); missing if person is censored or alive at study end date | numeric |
Note: R variable names are case sensitive! All variable names must be lower case. This can be done in any software. Below is an example person file format:
| id | gender | race | dob | pybegin | dlo | vs | rev | code |
|---|---|---|---|---|---|---|---|---|
| 1 | M | W | 11/20/1945 | 12/21/1970 | 7/31/2016 | |||
| 2 | M | N | 3/15/1942 | 1/19/1972 | 8/19/2011 | D | 10 | G30.9 |
| 3 | F | W | 6/5/1955 | 11/23/1970 | 12/7/1972 | D | 8 | 410.9 |
NOTE: Paths in R use forward slashes (/) instead of the standard back slashes () used by windows.
The date columns will need to be converted to R date values. This can be done using the as.Date() function. This function requires the user to specify the format of the dates being converted. Below gives a key in how to describe the date format:
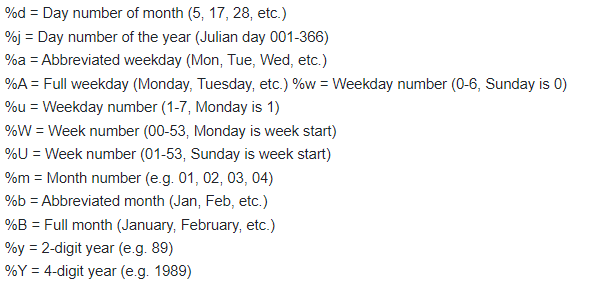
So, the above date columns in the above example person file have the
general format of “%m/%d/%Y”.
Below gives example code to read in the person file as well as code to
convert the date columns to date values:
library(LTASR)
library(tidyverse)
person <- read_csv('C:/person.csv') %>%
mutate(dob = as.Date(dob, format='%m/%d/%Y'),
pybegin = as.Date(pybegin, format='%m/%d/%Y'),
dlo = as.Date(dlo, format='%m/%d/%Y'))The LTASR package comes with an example person file, called person_example, that can be used for testing. For this example, it will be read in, its dates will be converted and will be saved as ‘person’ similar to the above code:
person <- person_example %>%
mutate(dob = as.Date(dob, format='%m/%d/%Y'),
pybegin = as.Date(pybegin, format='%m/%d/%Y'),
dlo = as.Date(dlo, format='%m/%d/%Y'))rateobj <- parseRate('C:/UCOD_WashingtonState012018.xml')Alternatively, the LTASR package comes pre-installed with a rate object for the 119 LTAS minors of the US population from 1960-2022. This can be used in any analyses, but must be explicitly called first:
rateobj <- us_119ucod_recentpy_table <- get_table(person, rateobj)Once the py_table has been created, it can be saved using the write_csv() function. This file contains person day counts (pdays) and outcome counts for each minor (_o1, _o2, …..).
With your stratified data, you can quickly create a table with of all SMRs for each minor found in the rate file using the smr_minor() function as below:
smr_minor_table <- smr_minor(py_table, rateobj)View(smr_minor_table)
write_csv(smr_minor_table, "C:/SMR_Minors.csv")smr_major_table <- smr_major(smr_minor_table, rateobj)
View(smr_major_table)
write_csv(smr_major_table, "C:/SMR_Majors")Additionally, any custom grouping can be calculated as well using the smr_custom() function where grouping are defined as a vector of numbers. Below defines groupings for all deaths and all cancers:
minor_grouping <- 1:119
all_causes <- smr_custom(smr_minor_table, minor_grouping)
View(all_causes)
minor_grouping <- 4:40
all_cancers <- smr_custom(smr_minor_table, minor_grouping)
View(all_cancers)These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.