
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
library("lavaan")## This is lavaan 0.6-8
## lavaan is FREE software! Please report any bugs.source("R/ICED_syntax.R")
source("R/ICED_run.R")
source("R/ICED_boot.R")
source("R/str2cov.R")
source("R/sim_ICED.R")
This script is intended to highlight progress and functionality with the ICED package
The ICED_syntax() function takes a dataframe and
generates the lavaan syntax to run the model (the first variable must be
time). Here we save it to the syn object. The model will
also be printed to the console. We are aiming to recreate the model from
Brandmaier et al. (2018), Figure 4
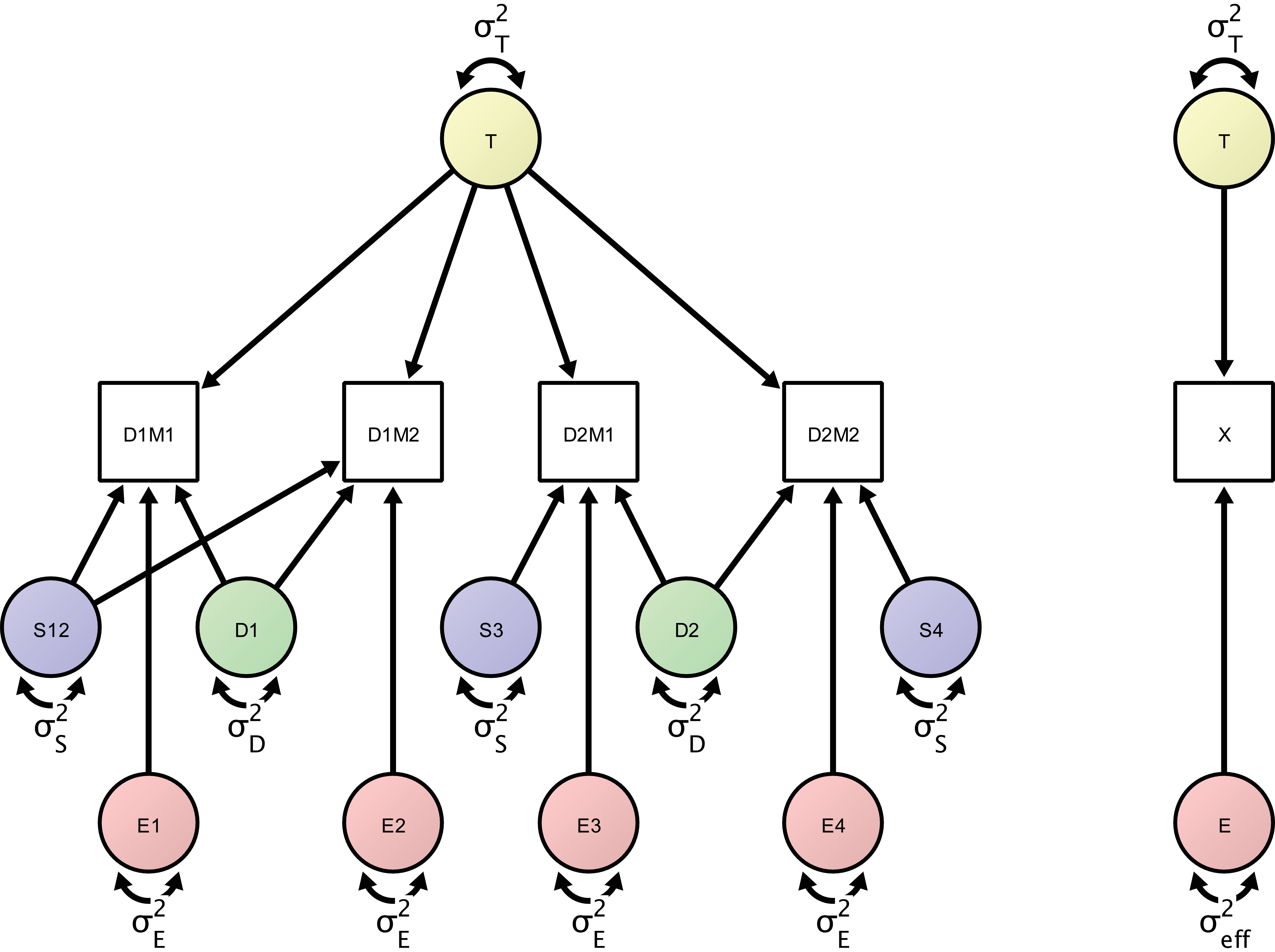
struc <- data.frame(time = c("T1", "T2", "T3", "T4"),
day = c("day1","day1","day2","day2"),
session = c("ses1", "ses1","ses2", "ses3"))
syn <- iced_syntax(struc)## ! regressions
## T =~ 1*T1
## T =~ 1*T2
## T =~ 1*T3
## T =~ 1*T4
## day1 =~ 1*T1
## day1 =~ 1*T2
## day2 =~ 1*T3
## day2 =~ 1*T4
## ses1 =~ 1*T1
## ses1 =~ 1*T2
## ses2 =~ 1*T3
## ses3 =~ 1*T4
## E1 =~ 1*T1
## E2 =~ 1*T2
## E3 =~ 1*T3
## E4 =~ 1*T4
## ! residuals, variances and covariances
## T ~~ time*T
## day1 ~~ day*day1
## day2 ~~ day*day2
## ses1 ~~ session*ses1
## ses2 ~~ session*ses2
## ses3 ~~ session*ses3
## E1 ~~ e*E1
## E2 ~~ e*E2
## E3 ~~ e*E3
## E4 ~~ e*E4
## T ~~ 0*day1
## T ~~ 0*day2
## T ~~ 0*ses1
## T ~~ 0*ses2
## T ~~ 0*ses3
## T ~~ 0*E1
## T ~~ 0*E2
## T ~~ 0*E3
## T ~~ 0*E4
## day1 ~~ 0*day2
## day1 ~~ 0*ses1
## day1 ~~ 0*ses2
## day1 ~~ 0*ses3
## day1 ~~ 0*E1
## day1 ~~ 0*E2
## day1 ~~ 0*E3
## day1 ~~ 0*E4
## day2 ~~ 0*ses1
## day2 ~~ 0*ses2
## day2 ~~ 0*ses3
## day2 ~~ 0*E1
## day2 ~~ 0*E2
## day2 ~~ 0*E3
## day2 ~~ 0*E4
## ses1 ~~ 0*ses2
## ses1 ~~ 0*ses3
## ses1 ~~ 0*E1
## ses1 ~~ 0*E2
## ses1 ~~ 0*E3
## ses1 ~~ 0*E4
## ses2 ~~ 0*ses3
## ses2 ~~ 0*E1
## ses2 ~~ 0*E2
## ses2 ~~ 0*E3
## ses2 ~~ 0*E4
## ses3 ~~ 0*E1
## ses3 ~~ 0*E2
## ses3 ~~ 0*E3
## ses3 ~~ 0*E4
## E1 ~~ 0*E2
## E1 ~~ 0*E3
## E1 ~~ 0*E4
## E2 ~~ 0*E3
## E2 ~~ 0*E4
## E3 ~~ 0*E4
## ! observed means
## T1~1
## T2~1
## T3~1
## T4~1
## !set lower bounds of variances
## time > 0.0001
## day > 0.0001
## session > 0.0001
## e > 0.0001We’ll simulate data to run the ICED model on. The
sim_ICED function takes the model structure dataframe we
used earlier and a list of variances for each latent variable. The
function returns several outputs, including the simulated data.
sim1 <- sim_ICED(struc,
variances = list(time = 10,
day = 2,
session = 1,
error = 3),
n = 2000)
head(sim1$data)## T1 T2 T3 T4
## 1 -6.892969387 -6.5932264 -8.723213 -7.6298887
## 2 2.563843435 6.7974728 1.521446 3.0070102
## 3 3.750682653 2.0135844 1.898953 3.3198217
## 4 -0.008140112 0.8534418 -2.378560 -0.9479120
## 5 -2.319533037 -2.1481638 -3.104065 -0.6225882
## 6 3.568513686 0.6051093 1.038575 -3.9176493we can also examine how well sim_ICED has recovered our
variance parameters by setting check_recovery = TRUE. lets
simulate two datasets, one large and another small.
sim2 <- sim_ICED(struc,
variances = list(time = 10,
day = 2,
session = 1,
error = 3),
n = 2000,
check_recovery = TRUE)## [1] "n = 2000 data simulated"
## [1] "data simulated based on ICC1 = 0.625"
## time day session error
## 10 2 1 3
## [1] "model parameters recovered:"
## [1] "ICC1 = 0.644583202724004"
## timeest dayest sessionest eest
## 10.5419826 1.9993960 0.8164775 2.9968714sim3 <- sim_ICED(struc,
variances = list(time = 10,
day = 2,
session = 1,
error = 3),
n = 20,
check_recovery = TRUE)## [1] "n = 20 data simulated"
## [1] "data simulated based on ICC1 = 0.625"
## time day session error
## 10 2 1 3
## [1] "model parameters recovered:"
## [1] "ICC1 = 0.540163701082888"
## timeest dayest sessionest eest
## 6.4965675 0.5116045 2.2645328 2.7543297The sim_ICED function uses a helper function
str2cov, which takes the same structure data.frame and the
list of variances we specified earlier to generate the expected
covariance of the model. This is then passed to mvrnorm to
generate the data. e.g.
str2cov(struc,
list(time = 10,
day = 2,
session = 1,
error = 3),
e_label = "error")## T1 T2 T3 T4
## T1 16 13 10 10
## T2 13 16 10 10
## T3 10 10 16 12
## T4 10 10 12 16we can now run our model. The run_ICED function will
print a bunch of relevant outputs. Note that the sim_ICED
function returns a list of objects, so we need to specify the data
part
res1 <- run_ICED(model = syn,
data = sim1$data)## $ICC
## [1] 0.6315415
##
## $ICC2
## [1] 0.828968
##
## $timeest
## [1] 10.12744
##
## $dayest
## [1] 2.023995
##
## $sessionest
## [1] 0.8988333
##
## $eest
## [1] 2.985794
##
## $EffectiveError
## [1] 2.089485
##
## $AbsoluteError
## [1] 2.058057
##
## $phi_dependability
## [1] 0.831106
##
## $lavaan
## lavaan 0.6-8 ended normally after 209 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 14
## Number of inequality constraints 4
##
## Number of observations 2000
## Number of missing patterns 1
##
## Model Test User Model:
##
## Test statistic 3.685
## Degrees of freedom 6
## P-value (Chi-square) 0.719
##
## $est_cov
## T1 T2 T3 T4
## T1 16.036
## T2 13.050 16.036
## T3 10.127 10.127 16.036
## T4 10.127 10.127 12.151 16.036we can also bootstrap our estimates. The output now includes 95% CIs on the ICC and ICC2. Best to use more than 10 boots, but set to 10 for speed here
run_ICED(model = syn,
data = sim1$data,
boot = 10)## Warning in norm.inter(t, alpha): extreme order statistics used as endpoints
## Warning in norm.inter(t, alpha): extreme order statistics used as endpoints
## $ICC
## [1] 0.6315415
##
## $ICC_CIs
## [1] 0.6190221 0.6469655
##
## $ICC2
## [1] 0.828968
##
## $ICC2_CIs
## [1] 0.8237431 0.8462589
##
## $timeest
## [1] 10.12744
##
## $dayest
## [1] 2.023995
##
## $sessionest
## [1] 0.8988333
##
## $eest
## [1] 2.985794
##
## $EffectiveError
## [1] 2.089485
##
## $AbsoluteError
## [1] 2.058057
##
## $phi_dependability
## [1] 0.831106
##
## $lavaan
## lavaan 0.6-8 ended normally after 209 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 14
## Number of inequality constraints 4
##
## Number of observations 2000
## Number of missing patterns 1
##
## Model Test User Model:
##
## Test statistic 3.685
## Degrees of freedom 6
## P-value (Chi-square) 0.719
##
## $est_cov
## T1 T2 T3 T4
## T1 16.036
## T2 13.050 16.036
## T3 10.127 10.127 16.036
## T4 10.127 10.127 12.151 16.036we can compare alternative models, for example constraining the variance of one component to zero
syntax2 <- iced_syntax(struc,
set_variances = c(res1$timeest,
res1$dayest,
0,
res1$eest))## Warning in iced_syntax(struc, set_variances = c(res1$timeest, res1$dayest, :
## set_variances must be list
## ! regressions
## T =~ 1*T1
## T =~ 1*T2
## T =~ 1*T3
## T =~ 1*T4
## day1 =~ 1*T1
## day1 =~ 1*T2
## day2 =~ 1*T3
## day2 =~ 1*T4
## ses1 =~ 1*T1
## ses1 =~ 1*T2
## ses2 =~ 1*T3
## ses3 =~ 1*T4
## E1 =~ 1*T1
## E2 =~ 1*T2
## E3 =~ 1*T3
## E4 =~ 1*T4
## ! residuals, variances and covariances
## T ~~ time*T
## day1 ~~ day*day1
## day2 ~~ day*day2
## ses1 ~~ session*ses1
## ses2 ~~ session*ses2
## ses3 ~~ session*ses3
## E1 ~~ e*E1
## E2 ~~ e*E2
## E3 ~~ e*E3
## E4 ~~ e*E4
## T ~~ 0*day1
## T ~~ 0*day2
## T ~~ 0*ses1
## T ~~ 0*ses2
## T ~~ 0*ses3
## T ~~ 0*E1
## T ~~ 0*E2
## T ~~ 0*E3
## T ~~ 0*E4
## day1 ~~ 0*day2
## day1 ~~ 0*ses1
## day1 ~~ 0*ses2
## day1 ~~ 0*ses3
## day1 ~~ 0*E1
## day1 ~~ 0*E2
## day1 ~~ 0*E3
## day1 ~~ 0*E4
## day2 ~~ 0*ses1
## day2 ~~ 0*ses2
## day2 ~~ 0*ses3
## day2 ~~ 0*E1
## day2 ~~ 0*E2
## day2 ~~ 0*E3
## day2 ~~ 0*E4
## ses1 ~~ 0*ses2
## ses1 ~~ 0*ses3
## ses1 ~~ 0*E1
## ses1 ~~ 0*E2
## ses1 ~~ 0*E3
## ses1 ~~ 0*E4
## ses2 ~~ 0*ses3
## ses2 ~~ 0*E1
## ses2 ~~ 0*E2
## ses2 ~~ 0*E3
## ses2 ~~ 0*E4
## ses3 ~~ 0*E1
## ses3 ~~ 0*E2
## ses3 ~~ 0*E3
## ses3 ~~ 0*E4
## E1 ~~ 0*E2
## E1 ~~ 0*E3
## E1 ~~ 0*E4
## E2 ~~ 0*E3
## E2 ~~ 0*E4
## E3 ~~ 0*E4
## ! observed means
## T1~1
## T2~1
## T3~1
## T4~1
## !set variances
##
## time == 10.1274398251025
## day == 2.02399517103763
## session == 0
## e == 2.98579433458071res2 <- run_ICED(syntax2,
sim1$data)## $ICC
## [1] 0.6690418
##
## $ICC2
## [1] 0.8520559
##
## $timeest
## [1] 10.12744
##
## $dayest
## [1] 2.023995
##
## $sessionest
## [1] 0
##
## $eest
## [1] 2.985794
##
## $EffectiveError
## [1] 1.758446
##
## $AbsoluteError
## [1] 1.758446
##
## $phi_dependability
## [1] 0.8520559
##
## $lavaan
## lavaan 0.6-8 ended normally after 1 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 14
## Number of equality constraints 10
##
## Number of observations 2000
## Number of missing patterns 1
##
## Model Test User Model:
##
## Test statistic 113.780
## Degrees of freedom 10
## P-value (Chi-square) 0.000
##
## $est_cov
## T1 T2 T3 T4
## T1 15.137
## T2 12.151 15.137
## T3 10.127 10.127 15.137
## T4 10.127 10.127 12.151 15.137anova(res1$lavaan,
res2$lavaan)## Chi-Squared Difference Test
##
## Df AIC BIC Chisq Chisq diff Df diff Pr(>Chisq)
## res1$lavaan 6 39651 39696 3.6852
## res2$lavaan 10 39753 39776 113.7797 110.09 4 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1we can use iced_syntax to generate the syntax for
multiple groups. The user can specify a number of groups or a vector of
strings. Here we’ll also highlight that we can set the variances to be
greater than zero also.
group_syntax <- iced_syntax(struc,
groups = 2,
fix_lower_bounds = TRUE)## ! regressions
## T =~ 1*T1
## T =~ 1*T2
## T =~ 1*T3
## T =~ 1*T4
## day1 =~ 1*T1
## day1 =~ 1*T2
## day2 =~ 1*T3
## day2 =~ 1*T4
## ses1 =~ 1*T1
## ses1 =~ 1*T2
## ses2 =~ 1*T3
## ses3 =~ 1*T4
## E1 =~ 1*T1
## E2 =~ 1*T2
## E3 =~ 1*T3
## E4 =~ 1*T4
## ! residuals, variances and covariances
## T ~~ c(lattime1,lattime2)*T
## day1 ~~ c(latday1,latday2)*day1
## day2 ~~ c(latday1,latday2)*day2
## ses1 ~~ c(latsession1,latsession2)*ses1
## ses2 ~~ c(latsession1,latsession2)*ses2
## ses3 ~~ c(latsession1,latsession2)*ses3
## E1 ~~ c(late1,late2)*E1
## E2 ~~ c(late1,late2)*E2
## E3 ~~ c(late1,late2)*E3
## E4 ~~ c(late1,late2)*E4
## T ~~ 0*day1
## T ~~ 0*day2
## T ~~ 0*ses1
## T ~~ 0*ses2
## T ~~ 0*ses3
## T ~~ 0*E1
## T ~~ 0*E2
## T ~~ 0*E3
## T ~~ 0*E4
## day1 ~~ 0*day2
## day1 ~~ 0*ses1
## day1 ~~ 0*ses2
## day1 ~~ 0*ses3
## day1 ~~ 0*E1
## day1 ~~ 0*E2
## day1 ~~ 0*E3
## day1 ~~ 0*E4
## day2 ~~ 0*ses1
## day2 ~~ 0*ses2
## day2 ~~ 0*ses3
## day2 ~~ 0*E1
## day2 ~~ 0*E2
## day2 ~~ 0*E3
## day2 ~~ 0*E4
## ses1 ~~ 0*ses2
## ses1 ~~ 0*ses3
## ses1 ~~ 0*E1
## ses1 ~~ 0*E2
## ses1 ~~ 0*E3
## ses1 ~~ 0*E4
## ses2 ~~ 0*ses3
## ses2 ~~ 0*E1
## ses2 ~~ 0*E2
## ses2 ~~ 0*E3
## ses2 ~~ 0*E4
## ses3 ~~ 0*E1
## ses3 ~~ 0*E2
## ses3 ~~ 0*E3
## ses3 ~~ 0*E4
## E1 ~~ 0*E2
## E1 ~~ 0*E3
## E1 ~~ 0*E4
## E2 ~~ 0*E3
## E2 ~~ 0*E4
## E3 ~~ 0*E4
## ! observed means
## T1~1
## T2~1
## T3~1
## T4~1
## !set lower bounds of variances
## lattime1 > 0.0001
## lattime2 > 0.0001
## latday1 > 0.0001
## latday2 > 0.0001
## latsession1 > 0.0001
## latsession2 > 0.0001
## late1 > 0.0001
## late2 > 0.0001group_syntax <- iced_syntax(struc,
groups = c("group1", "group2"),
fix_lower_bounds = TRUE)## ! regressions
## T =~ 1*T1
## T =~ 1*T2
## T =~ 1*T3
## T =~ 1*T4
## day1 =~ 1*T1
## day1 =~ 1*T2
## day2 =~ 1*T3
## day2 =~ 1*T4
## ses1 =~ 1*T1
## ses1 =~ 1*T2
## ses2 =~ 1*T3
## ses3 =~ 1*T4
## E1 =~ 1*T1
## E2 =~ 1*T2
## E3 =~ 1*T3
## E4 =~ 1*T4
## ! residuals, variances and covariances
## T ~~ c(lattimegroup1,lattimegroup2)*T
## day1 ~~ c(latdaygroup1,latdaygroup2)*day1
## day2 ~~ c(latdaygroup1,latdaygroup2)*day2
## ses1 ~~ c(latsessiongroup1,latsessiongroup2)*ses1
## ses2 ~~ c(latsessiongroup1,latsessiongroup2)*ses2
## ses3 ~~ c(latsessiongroup1,latsessiongroup2)*ses3
## E1 ~~ c(lategroup1,lategroup2)*E1
## E2 ~~ c(lategroup1,lategroup2)*E2
## E3 ~~ c(lategroup1,lategroup2)*E3
## E4 ~~ c(lategroup1,lategroup2)*E4
## T ~~ 0*day1
## T ~~ 0*day2
## T ~~ 0*ses1
## T ~~ 0*ses2
## T ~~ 0*ses3
## T ~~ 0*E1
## T ~~ 0*E2
## T ~~ 0*E3
## T ~~ 0*E4
## day1 ~~ 0*day2
## day1 ~~ 0*ses1
## day1 ~~ 0*ses2
## day1 ~~ 0*ses3
## day1 ~~ 0*E1
## day1 ~~ 0*E2
## day1 ~~ 0*E3
## day1 ~~ 0*E4
## day2 ~~ 0*ses1
## day2 ~~ 0*ses2
## day2 ~~ 0*ses3
## day2 ~~ 0*E1
## day2 ~~ 0*E2
## day2 ~~ 0*E3
## day2 ~~ 0*E4
## ses1 ~~ 0*ses2
## ses1 ~~ 0*ses3
## ses1 ~~ 0*E1
## ses1 ~~ 0*E2
## ses1 ~~ 0*E3
## ses1 ~~ 0*E4
## ses2 ~~ 0*ses3
## ses2 ~~ 0*E1
## ses2 ~~ 0*E2
## ses2 ~~ 0*E3
## ses2 ~~ 0*E4
## ses3 ~~ 0*E1
## ses3 ~~ 0*E2
## ses3 ~~ 0*E3
## ses3 ~~ 0*E4
## E1 ~~ 0*E2
## E1 ~~ 0*E3
## E1 ~~ 0*E4
## E2 ~~ 0*E3
## E2 ~~ 0*E4
## E3 ~~ 0*E4
## ! observed means
## T1~1
## T2~1
## T3~1
## T4~1
## !set lower bounds of variances
## lattimegroup1 > 0.0001
## lattimegroup2 > 0.0001
## latdaygroup1 > 0.0001
## latdaygroup2 > 0.0001
## latsessiongroup1 > 0.0001
## latsessiongroup2 > 0.0001
## lategroup1 > 0.0001
## lategroup2 > 0.0001generating multiple group data takes a few more lines of code currently - but I plan to adapt this to take vectors within the lists.
variances_hi <- list(time = 8,
day = .25,
session = .2,
error = .25)
variances_lo <- list(time = 2,
day = .25,
session = .2,
error = .25)
sim_hi <- sim_ICED(structure = struc,
variances = variances_hi,
n = 100)$data
sim_lo <- sim_ICED(structure = struc,
variances = variances_lo,
n = 100)$data
sim_hi$group <- "high"
sim_lo$group <- "low"
sim_all <- rbind(sim_hi, sim_lo)we can check the ICC reliability we have specified fairly easily,
e.g. for the high group
variances_hi$time / (sum(unlist(variances_hi))). For the
high group ICC = 0.9195402, and the low group ICC = 0.7407407.
Then we can compare the groups (not currenly within run_ICED). Here, m0 is our base model constraining variances across groups, and m1 using the model we just generated to allow them to vary across groups.
m1 <- lavaan::lavaan(model = group_syntax,
data = sim_all,
group = "group")
m0 <- lavaan::lavaan(model = syn,
data = sim_all,
group = "group")## Warning in lavaanify(model = FLAT, constraints = constraints, varTable = lavdata@ov, : lavaan WARNING: using a single label per parameter in a multiple group
## setting implies imposing equality constraints across all the groups;
## If this is not intended, either remove the label(s), or use a vector
## of labels (one for each group);
## See the Multiple groups section in the man page of model.syntax.summary(m1)## lavaan 0.6-8 ended normally after 376 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 28
## Number of inequality constraints 8
##
## Number of observations per group:
## high 100
## low 100
##
## Model Test User Model:
##
## Test statistic 15.425
## Degrees of freedom 12
## P-value (Chi-square) 0.219
## Test statistic for each group:
## high 7.306
## low 8.119
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
##
## Group 1 [high]:
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## T =~
## T1 1.000
## T2 1.000
## T3 1.000
## T4 1.000
## day1 =~
## T1 1.000
## T2 1.000
## day2 =~
## T3 1.000
## T4 1.000
## ses1 =~
## T1 1.000
## T2 1.000
## ses2 =~
## T3 1.000
## ses3 =~
## T4 1.000
## E1 =~
## T1 1.000
## E2 =~
## T2 1.000
## E3 =~
## T3 1.000
## E4 =~
## T4 1.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## T ~~
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day1 ~~
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day2 ~~
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses1 ~~
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses2 ~~
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses3 ~~
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## E1 ~~
## E2 0.000
## E3 0.000
## E4 0.000
## E2 ~~
## E3 0.000
## E4 0.000
## E3 ~~
## E4 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .T1 -0.013 0.296 -0.044 0.965
## .T2 0.043 0.296 0.146 0.884
## .T3 0.122 0.296 0.411 0.681
## .T4 0.067 0.296 0.227 0.821
## T 0.000
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## T (ltt1) 7.985 1.174 6.798 0.000
## day1 (ltd1) 0.407 0.098 4.172 0.000
## day2 (ltd1) 0.407 0.098 4.172 0.000
## ses1 (lts1) 0.153 0.061 2.535 0.011
## ses2 (lts1) 0.153 0.061 2.535 0.011
## ses3 (lts1) 0.153 0.061 2.535 0.011
## E1 (ltg1) 0.216 0.031 7.073 0.000
## E2 (ltg1) 0.216 0.031 7.073 0.000
## E3 (ltg1) 0.216 0.031 7.073 0.000
## E4 (ltg1) 0.216 0.031 7.073 0.000
## .T1 0.000
## .T2 0.000
## .T3 0.000
## .T4 0.000
##
##
## Group 2 [low]:
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## T =~
## T1 1.000
## T2 1.000
## T3 1.000
## T4 1.000
## day1 =~
## T1 1.000
## T2 1.000
## day2 =~
## T3 1.000
## T4 1.000
## ses1 =~
## T1 1.000
## T2 1.000
## ses2 =~
## T3 1.000
## ses3 =~
## T4 1.000
## E1 =~
## T1 1.000
## E2 =~
## T2 1.000
## E3 =~
## T3 1.000
## E4 =~
## T4 1.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## T ~~
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day1 ~~
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day2 ~~
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses1 ~~
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses2 ~~
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses3 ~~
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## E1 ~~
## E2 0.000
## E3 0.000
## E4 0.000
## E2 ~~
## E3 0.000
## E4 0.000
## E3 ~~
## E4 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .T1 0.056 0.180 0.310 0.757
## .T2 0.038 0.180 0.212 0.832
## .T3 0.035 0.180 0.191 0.848
## .T4 0.038 0.180 0.210 0.833
## T 0.000
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## T (ltt2) 2.639 0.407 6.488 0.000
## day1 (ltd2) 0.243 0.076 3.203 0.001
## day2 (ltd2) 0.243 0.076 3.203 0.001
## ses1 (lts2) 0.119 0.063 1.905 0.057
## ses2 (lts2) 0.119 0.063 1.905 0.057
## ses3 (lts2) 0.119 0.063 1.905 0.057
## E1 (ltg2) 0.250 0.035 7.081 0.000
## E2 (ltg2) 0.250 0.035 7.081 0.000
## E3 (ltg2) 0.250 0.035 7.081 0.000
## E4 (ltg2) 0.250 0.035 7.081 0.000
## .T1 0.000
## .T2 0.000
## .T3 0.000
## .T4 0.000
##
## Constraints:
## |Slack|
## lattimegroup1 - (0.0001) 7.985
## lattimegroup2 - (0.0001) 2.639
## latdaygroup1 - (0.0001) 0.407
## latdaygroup2 - (0.0001) 0.243
## latsessiongroup1 - (0.0001) 0.153
## latsessiongroup2 - (0.0001) 0.119
## lategroup1 - (0.0001) 0.216
## lategroup2 - (0.0001) 0.249summary(m0)## lavaan 0.6-8 ended normally after 434 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 28
## Number of inequality constraints 4
##
## Number of observations per group:
## high 100
## low 100
##
## Model Test User Model:
##
## Test statistic 45.492
## Degrees of freedom 16
## P-value (Chi-square) 0.000
## Test statistic for each group:
## high 17.723
## low 27.769
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
##
## Group 1 [high]:
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## T =~
## T1 1.000
## T2 1.000
## T3 1.000
## T4 1.000
## day1 =~
## T1 1.000
## T2 1.000
## day2 =~
## T3 1.000
## T4 1.000
## ses1 =~
## T1 1.000
## T2 1.000
## ses2 =~
## T3 1.000
## ses3 =~
## T4 1.000
## E1 =~
## T1 1.000
## E2 =~
## T2 1.000
## E3 =~
## T3 1.000
## E4 =~
## T4 1.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## T ~~
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day1 ~~
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day2 ~~
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses1 ~~
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses2 ~~
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses3 ~~
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## E1 ~~
## E2 0.000
## E3 0.000
## E4 0.000
## E2 ~~
## E3 0.000
## E4 0.000
## E3 ~~
## E4 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .T1 -0.013 0.245 -0.053 0.957
## .T2 0.043 0.245 0.176 0.860
## .T3 0.122 0.245 0.496 0.620
## .T4 0.067 0.245 0.274 0.784
## T 0.000
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## T (time) 5.312 0.559 9.503 0.000
## day1 (day) 0.326 0.061 5.327 0.000
## day2 (day) 0.326 0.061 5.327 0.000
## ses1 (sssn) 0.135 0.044 3.104 0.002
## ses2 (sssn) 0.135 0.044 3.104 0.002
## ses3 (sssn) 0.135 0.044 3.104 0.002
## E1 (e) 0.233 0.023 10.006 0.000
## E2 (e) 0.233 0.023 10.006 0.000
## E3 (e) 0.233 0.023 10.006 0.000
## E4 (e) 0.233 0.023 10.006 0.000
## .T1 0.000
## .T2 0.000
## .T3 0.000
## .T4 0.000
##
##
## Group 2 [low]:
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## T =~
## T1 1.000
## T2 1.000
## T3 1.000
## T4 1.000
## day1 =~
## T1 1.000
## T2 1.000
## day2 =~
## T3 1.000
## T4 1.000
## ses1 =~
## T1 1.000
## T2 1.000
## ses2 =~
## T3 1.000
## ses3 =~
## T4 1.000
## E1 =~
## T1 1.000
## E2 =~
## T2 1.000
## E3 =~
## T3 1.000
## E4 =~
## T4 1.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## T ~~
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day1 ~~
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## day2 ~~
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses1 ~~
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses2 ~~
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## ses3 ~~
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
## E1 ~~
## E2 0.000
## E3 0.000
## E4 0.000
## E2 ~~
## E3 0.000
## E4 0.000
## E3 ~~
## E4 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .T1 0.056 0.245 0.228 0.820
## .T2 0.038 0.245 0.156 0.876
## .T3 0.035 0.245 0.141 0.888
## .T4 0.038 0.245 0.155 0.877
## T 0.000
## day1 0.000
## day2 0.000
## ses1 0.000
## ses2 0.000
## ses3 0.000
## E1 0.000
## E2 0.000
## E3 0.000
## E4 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## T (time) 5.312 0.559 9.503 0.000
## day1 (day) 0.326 0.061 5.327 0.000
## day2 (day) 0.326 0.061 5.327 0.000
## ses1 (sssn) 0.135 0.044 3.104 0.002
## ses2 (sssn) 0.135 0.044 3.104 0.002
## ses3 (sssn) 0.135 0.044 3.104 0.002
## E1 (e) 0.233 0.023 10.006 0.000
## E2 (e) 0.233 0.023 10.006 0.000
## E3 (e) 0.233 0.023 10.006 0.000
## E4 (e) 0.233 0.023 10.006 0.000
## .T1 0.000
## .T2 0.000
## .T3 0.000
## .T4 0.000
##
## Constraints:
## |Slack|
## time - (0.0001) 5.312
## day - (0.0001) 0.326
## session - (0.0001) 0.135
## e - (0.0001) 0.233anova(m1, m0)## Chi-Squared Difference Test
##
## Df AIC BIC Chisq Chisq diff Df diff Pr(>Chisq)
## m1 12 2419.8 2472.5 15.425
## m0 16 2441.8 2481.4 45.492 30.067 4 4.743e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.