
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

EstimateBreed is an R package designed to perform analyses and estimate environmental covariates and genetic parameters related to selection strategies and the development of superior genotypes. The package offers two main functionalities:
You can install the development version of EstimateBreed from GitHub with:
if (!requireNamespace("pak", quietly = TRUE)) {
install.packages("pak")
}
pak::pak("willyanjnr/EstimateBreed")Obtain the genetic selection index for resilience (ISGR) for selecting genotypes for environmental stressors, as described by Bandeira et al. (2024).
library(EstimateBreed)
#Obtain environmental deviations
data("desvamb")
DPclim <- with(desvamb,desv_clim(ENV,TMED,PREC))
DPclim
# A tibble: 3 × 5
# ENV STMED TMEDR SPREC PRECIR
# <chr> <dbl> <dbl> <dbl> <dbl>
# 1 E1 2.65 24.8 5.46 339.
# 2 E2 3.65 23.8 5.27 344.
# 3 E3 2.81 24.5 5.47 362.
#Get the ISGR
data("genot")
isgr_index <- with(genot, isgr(GEN,ENV,NG,MG,CICLO))
isgr_index
# Gen Env ISGR
# 26 L454 E1 6.489941
# 22 L455 E1 7.084315
# 19 L541 E1 7.653157
# 18 L367 E1 7.862185
# 16 L380 E1 8.329434
# 12 L393 E1 9.638909
# 10 L439 E1 10.552056
# 28 L298 E3 12.209433
# 30 L358 E2 23.347984
# 29 L346 E2 23.793351
# 27 L195 E2 24.719927
# 25 L179 E2 25.747317
# 24 L359 E2 26.300686
# 23 L345 E2 26.886419
# 1 L445 E1 27.255375
# 21 L185 E2 28.211433
# 20 L310 E2 28.942165
# 17 L178 E2 31.418785
# 15 L261 E2 33.424611
# 14 L269 E2 34.605133
# 13 L209 E2 35.959423
# 11 L263 E2 39.127798
# 9 L201 E2 43.145922
# 8 L299 E2 45.686042
# 7 L152 E2 48.926278
# 6 L26 E2 52.988109
# 5 L166 E2 57.596139
# 4 L155 E2 64.251152
# 3 L277 E2 74.756384
# 2 L162 E2 86.543916Selection of transgressive genotypes with the selection differential (mean and standard deviations).
library(EstimateBreed)
Gen <- paste0("G", 1:20)
Var <- round(rnorm(20, mean = 3.5, sd = 0.8), 2)
Control <- rep(3.8, 20)
data <- data.frame(Gen,Var,Control)
with(data,transg(Gen,Var,Control))Returns the general parameters and the genotypes selected for each treshold. Also plot a representative graph of the selected genotypes based on the mean and standard deviations.
---------------------------------------------------------------------
Selection of Transgressive Genotypes - Selection Differential (SD)
---------------------------------------------------------------------
Parameters:
---------------------------------------------------------------------
Overall Mean : 3.566
Control Mean : 3.800
Standard Deviation : 0.603
Mean + 1SD : 4.169
Mean + 2SD : 4.771
Mean + 3SD : 5.374
---------------------------------------------------------------------
Genotypes above each threshold:
---------------------------------------------------------------------
Genotypes above Control Mean : G4, G7, G8, G9, G12, G14,
G20
Genotype above Overall Mean : G4, G7, G8, G9, G12, G14,
G16, G18, G20
Genotypes above Mean + 1SD : G7, G9, G20
Genotypes above Mean + 2SD : G7
Genotypes above Mean + 3SD : None
---------------------------------------------------------------------Predict ∆T to determine the ideal times to apply agricultural pesticides.
library(EstimateBreed)
# Forecasting application conditions
tdelta(-53.696944444444,-28.063888888889,type=1,days=10)
# Retrospective analysis of application conditions
tdelta(-53.6969,-28.0638,type=2,days=10,dates=c("2023-01-01","2023-05-01"))Estimation of soybean plastochron using average air temperature and number of nodes
library(EstimateBreed)
data("pheno")
with(pheno, plast(GEN,TMED,EST,NN,habit="ind",plot=TRUE))#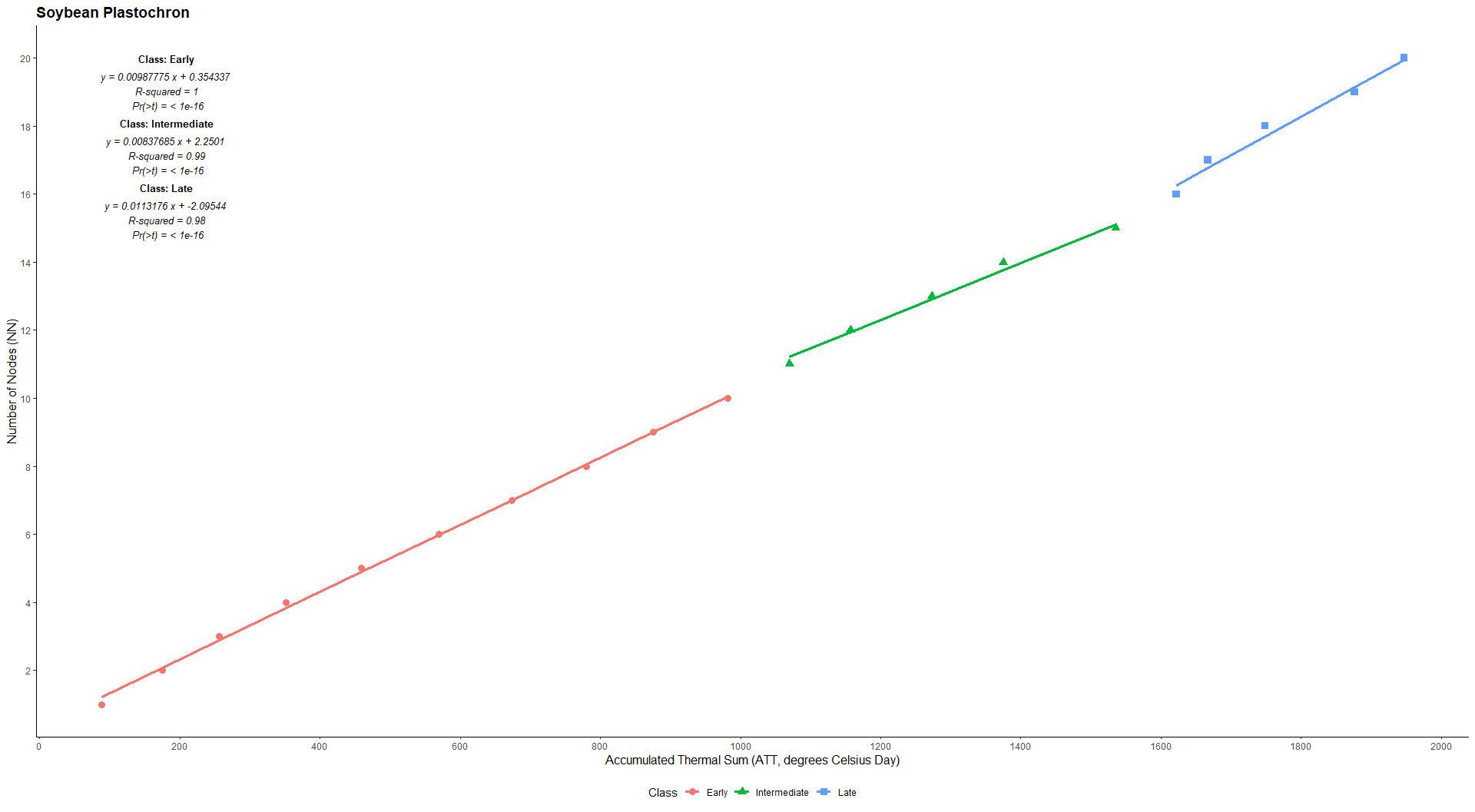
Complete documentation can be found when using the package within R.
When citing this package, please use,
library(EstimateBreed)
citation("EstimateBreed")
To cite package ‘EstimateBreed’ in publications use:
Willyan Jr. A. Bandeira, Ivan R. Carvalho, Murilo V. Loro, Leonardo
C. Pradebon, José A. G. da Silva (2025). _EstimateBreed: Estimation
of Environmental Variables and Genetic Parameters_. R package
version 0.1.0, <https://github.com/willyanjnr/EstimateBreed>.
A BibTeX entry for LaTeX users is
@Manual{,
title = {EstimateBreed: Estimation of Environmental Variables and Genetic Parameters},
author = {{Willyan Jr. A. Bandeira} and {Ivan R. Carvalho} and {Murilo V. Loro} and {Leonardo C. Pradebon} and {José A. G. da Silva}},
year = {2025},
note = {R package version 0.1.0},
url = {https://github.com/willyanjnr/EstimateBreed},
}These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.