The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This is a flexible R package for enrichment analysis based on user-defined sets. It allows users to perform over-representation analysis of the custom sets among any specified ranked feature list, hence making enrichment analysis applicable to various types of data from different scientific fields. EnrichIntersect also enables an interactive means to visualize identified associations based on, for example, the mix-lasso model (Zhao et al., 2022) or similar methods.
Install the latest released version from CRAN
install.packages("EnrichIntersect")
library("EnrichIntersect")Install the latest development version from GitHub
#install.packages("remotes")
remotes::install_github("ocbe-uio/EnrichIntersect")
library("EnrichIntersect")Data set cancers_drug_groups is a list including a score
dataframe with 147 drugs as rows and 19 cancer types as columns, and a
dataframe with nice pre-defined drug groups (1st column) of the 147
drugs (2nd column).
data(cancers_drug_groups, package = "EnrichIntersect")
x <- cancers_drug_groups$score
custom.set <- cancers_drug_groups$custom.set
set.seed(123)
enrich <- enrichment(x, custom.set, permute.n = 5)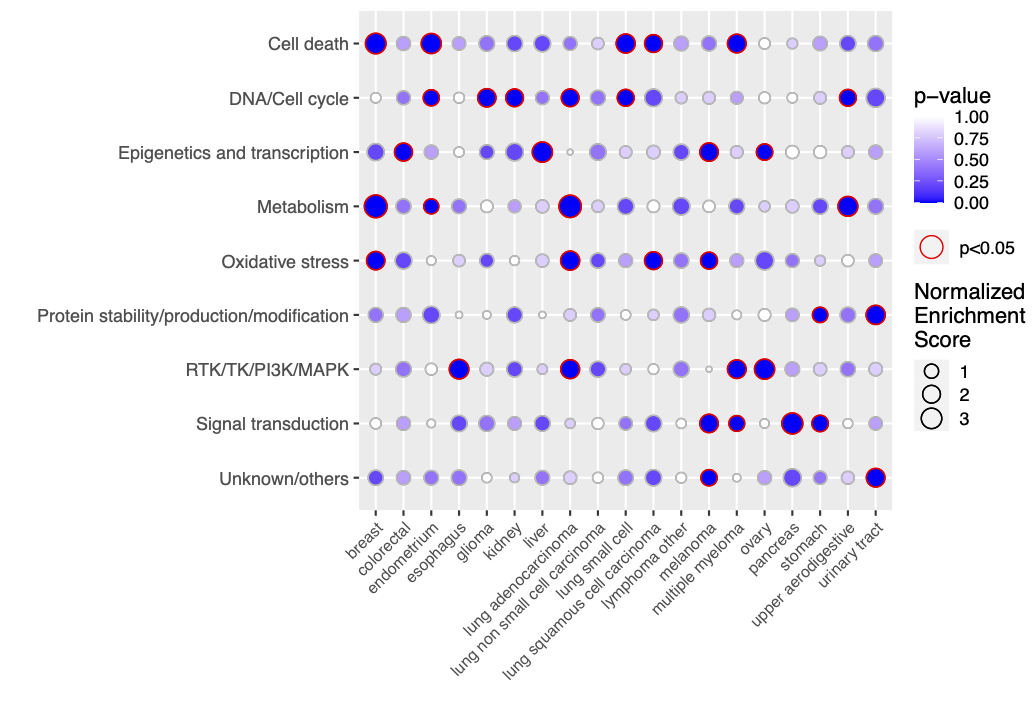
Data set cancers_genes_drugs is an array with
association scores between 56 genes (1st dimension), three cancer types
(2nd dimension) and two drugs (3rd dimension).
data(cancers_genes_drugs, package = "EnrichIntersect")
# better to use argument `out.fig = "pdf"` for printing a pdf or html figure
intersectSankey(cancers_genes_drugs, step.names = c("Cancers","Genes","Drugs"))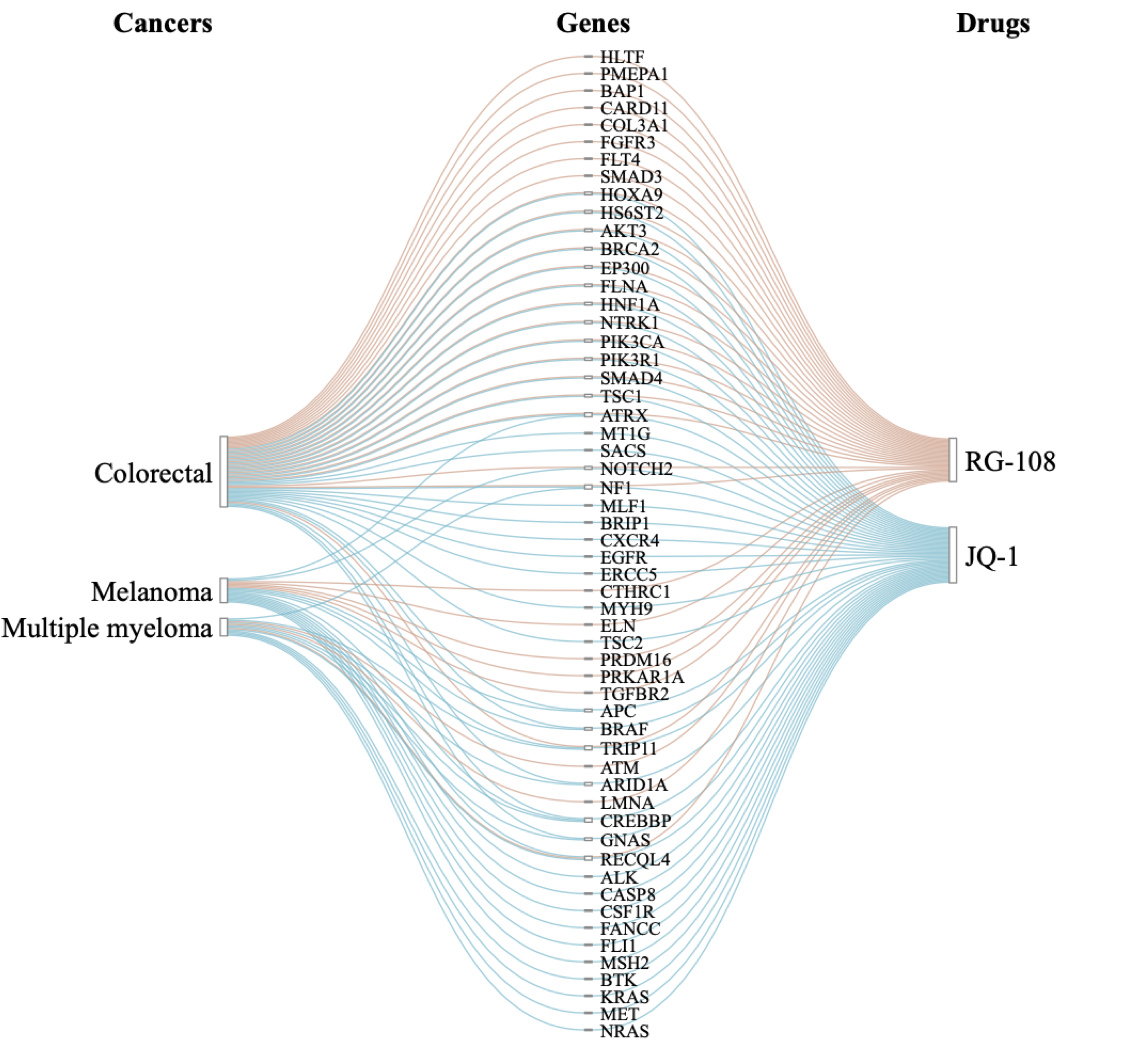
Zhi Zhao, Manuela Zucknick, Tero Aittokallio (2022). EnrichIntersect: an R package for custom set enrichment analysis and interactive visualization of intersecting sets. Bioinformatics Advances, vbac073. DOI: 10.1093/bioadv/vbac073.
Zhi Zhao, Shixiong Wang, Manuela Zucknick, Tero Aittokallio (2022). Tissue-specific identification of multi-omics features for pan-cancer drug response prediction. iScience, 25(8): 104767. DOI: 10.1016/j.isci.2022.104767.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.